| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
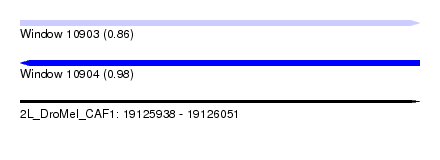
| Location | 19,125,938 – 19,126,051 |
| Length | 113 |
| Max. P | 0.984311 |

| Location | 19,125,938 – 19,126,051 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -22.28 |
| Energy contribution | -21.56 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
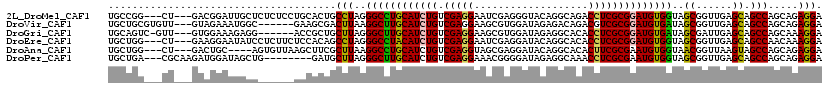
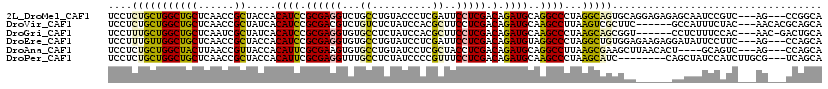
>2L_DroMel_CAF1 19125938 113 + 22407834 UGCCGG---CU---GACGGAUUGCUCUCUCCUGCACUGCCUAGGGCCUGCAUCUGUCGAGGAAUCGAGGGUACAGGCAGACCUCGCGGAUGUGGUAGCGGUUGAGCAGCCAGCAGAGGA (((.((---((---(...(((((((....((.(((((((..((((((((...((.((((....)))).))..)))))...))).)))).))))).)))))))...))))).)))..... ( -46.00) >DroVir_CAF1 14916 110 + 1 UGCUGCGUGUU---GUAGAAAUGGC------GAAGCGACUUAAGGCUUGCAUCUGUCGAGGAAGCGUGGAUAGAGACAGACGUCGCGGAUGUGAUAGCGGUUGAGCAGCCAGCAGAGGA ..((((..(((---((...(((.((------.((((........))))(((((((.(((.(...(.((........))).).))))))))))....)).)))..)))))..)))).... ( -32.70) >DroGri_CAF1 14398 109 + 1 UGCAGUC-GUU---GUGGAAAGAGG------ACCGCUGCUUAGGGCUUGCAUCUGUCGAGGAAGCGUGGAUAGAGGCACACCUCGCGGAUGUGAUAGCGAUUGAGCAGCCAGCAAAGGA (((.(((-.((---.......)).)------)).(((((((((.(((.(((((((.(((((..((.(......).))...))))))))))))...)))..)))))))))..)))..... ( -42.30) >DroEre_CAF1 12023 113 + 1 UGCUGG---CU---GAAGGAAUAUCCUCUUCUCCACAGCCUAGGGCCUACAUCUGUCGAGGAAUCGAGGAUACAGGCACACCUCGCGGAUGUGGUAGCGGUUGAGCAGCCAACAAAGGA ((.(((---((---(.(((.....)))........(((((....(((.(((((((.(((((...................)))))))))))))))...)))))..)))))).))..... ( -39.71) >DroAna_CAF1 12026 109 + 1 UGCUGG---CU---GACUGC----AGUGUUAAGCUUCGCUUAAGGCCUGCAUCUGUCGAGGUAGCGAGGAUACAGGCACACUUCGCGAAUGUGGUAACGGUUAAGUAGCCAGCAGAGGA ((((((---((---(.((..----(.(((((.(((((((.....(((((.((((.(((......))))))).))))).......))))).))..))))).)..)))))))))))..... ( -42.40) >DroPer_CAF1 18480 108 + 1 UGCUGA---CGCAAGAUGGAUAGCUG--------GAUGCUUAGGGCUUGCAUCUGUCGAGGAAACGGGGAUAGAGGCAAACCUCGCGAAUGUGGUAGCGGUUGAGCAGCCAGCAGAGGA (((...---.))).........((((--------(.(((((((((.((((.((((((..(....)...)))))).)))).))((((..........))))))))))).)))))...... ( -39.50) >consensus UGCUGG___CU___GAAGGAAUGCC______ACCGCUGCUUAGGGCCUGCAUCUGUCGAGGAAGCGAGGAUACAGGCACACCUCGCGGAUGUGGUAGCGGUUGAGCAGCCAGCAGAGGA ......................................(((..((((((((((((.(((((...................))))))))))))))..((......)).))).....))). (-22.28 = -21.56 + -0.72)
| Location | 19,125,938 – 19,126,051 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.28 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
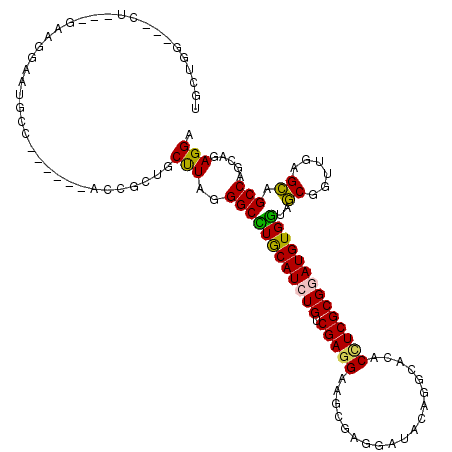
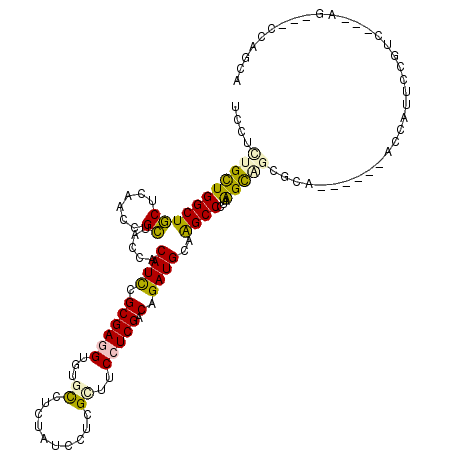
>2L_DroMel_CAF1 19125938 113 - 22407834 UCCUCUGCUGGCUGCUCAACCGCUACCACAUCCGCGAGGUCUGCCUGUACCCUCGAUUCCUCGACAGAUGCAGGCCCUAGGCAGUGCAGGAGAGAGCAAUCCGUC---AG---CCGGCA ......((((((((.......(((......((((((..((((((((((((..((((....))))..).)))))))...))))..))).)))...))).......)---))---))))). ( -43.84) >DroVir_CAF1 14916 110 - 1 UCCUCUGCUGGCUGCUCAACCGCUAUCACAUCCGCGACGUCUGUCUCUAUCCACGCUUCCUCGACAGAUGCAAGCCUUAAGUCGCUUC------GCCAUUUCUAC---AACACGCAGCA ....((((((((.((......))..........((((((((((((.................)))))))(....).....)))))...------)))).......---.....)))).. ( -23.03) >DroGri_CAF1 14398 109 - 1 UCCUUUGCUGGCUGCUCAAUCGCUAUCACAUCCGCGAGGUGUGCCUCUAUCCACGCUUCCUCGACAGAUGCAAGCCCUAAGCAGCGGU------CCUCUUUCCAC---AAC-GACUGCA .....((((((((((((..((((..........))((((((((........))))))))...))..)).)).))))...))))(((((------(..........---...-)))))). ( -30.92) >DroEre_CAF1 12023 113 - 1 UCCUUUGUUGGCUGCUCAACCGCUACCACAUCCGCGAGGUGUGCCUGUAUCCUCGAUUCCUCGACAGAUGUAGGCCCUAGGCUGUGGAGAAGAGGAUAUUCCUUC---AG---CCAGCA .....(((((((((.....((((..(((((.((....)))))(((((((((.((((....))))..)))))))))....))..))))....((((.....)))))---))---)))))) ( -44.30) >DroAna_CAF1 12026 109 - 1 UCCUCUGCUGGCUACUUAACCGUUACCACAUUCGCGAAGUGUGCCUGUAUCCUCGCUACCUCGACAGAUGCAGGCCUUAAGCGAAGCUUAACACU----GCAGUC---AG---CCAGCA ......((((((((((.....((((.....(((((.....(.(((((((((.(((......)))..))))))))))....)))))...))))...----..))).---))---))))). ( -39.80) >DroPer_CAF1 18480 108 - 1 UCCUCUGCUGGCUGCUCAACCGCUACCACAUUCGCGAGGUUUGCCUCUAUCCCCGUUUCCUCGACAGAUGCAAGCCCUAAGCAUC--------CAGCUAUCCAUCUUGCG---UCAGCA ......(((((.((((....(((..........))).(((((((.(((.((...........)).))).)))))))...)))).)--------)))).........(((.---...))) ( -30.00) >consensus UCCUCUGCUGGCUGCUCAACCGCUACCACAUCCGCGAGGUGUGCCUCUAUCCUCGCUUCCUCGACAGAUGCAAGCCCUAAGCAGCGCA______ACCAUUCCGUC___AG___CCAGCA ....(((((((((((......)).....((((.((((((...((..........))..))))).).))))..))))...)))))................................... (-16.61 = -16.28 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:00 2006