| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,123,871 – 19,123,979 |
| Length | 108 |
| Max. P | 0.658282 |

| Location | 19,123,871 – 19,123,979 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 65.18 |
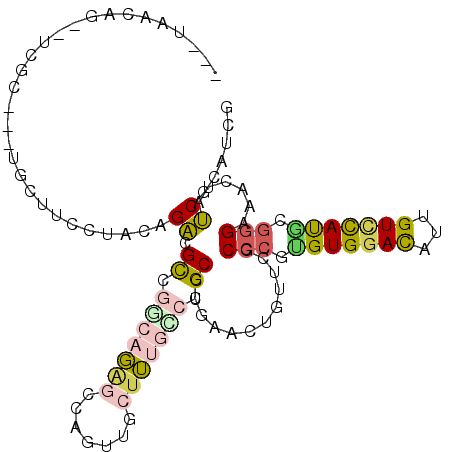
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19123871 108 + 22407834 AUGUAAAAGCAUCGUA--UUUUUCCCACAGACGUCGCCACAGCCAGUUGCUGGCGCUGCCCGAAUUGUUCACCCACGUGGACACUGUCUAUGCGGGCACGUAUCGCAUCG ........(((((((.--..............(.((((((((....))).)))))).(((((...(((((((....))))))).........)))))))).)).)).... ( -28.60) >DroVir_CAF1 11994 102 + 1 ---UUGCA---AUGCC--UUUGCUCUGCAGACGCCCACAGGCGCAAUUGCUUUGCCUGCCGGAGCUGUUCGCACGCGUGGACAUUGUGCACGGAGGACACUAUCGAAUCG ---.((((---.....--...(((((((((..(((....)))((((.....))))))).))))))(((((((....)))))))...))))(((((....)).)))..... ( -33.00) >DroPse_CAF1 17056 99 + 1 ---UAACAG-----C---CGCUUCUUGCAGGCGCGGGCAGAGUCAGCUCCUUUGCCUGCCCGAGCUGUUUGCCCGUGUGGACCUUGUCCAUACGGGAAACUACCGCAUCG ---.(((((-----(---(((.....)).((.((((((((((.......))))))))))))).))))))..(((((((((((...))))))))))).............. ( -47.90) >DroSec_CAF1 11016 107 + 1 -UGUUAAAGCAUCGUG--UUUUUCCCACAGACGUCGUCACAGCCAGUUGCUGGCGCUGCCCGAAUUGUUCACCCAUGUGGACACGGUCUACGCGGGCACAUAUCGUAUCG -......(((...(((--.......))).(((...)))...(((((...))))))))(((((...(((((((....)))))))((.....)))))))............. ( -30.00) >DroWil_CAF1 14905 104 + 1 -CAUUA-----UCAUUUUUGCUUCUUACAGACGUCCUCAGUCACAAUUGCUUUGCCUGCCUGAACUUUUUGCCCGUGUGGAUAUUGUUCAUUCGGGCAAUUAUCGCUUGG -.....-----................((..((...((((.(((((.....)))..)).)))).....(((((((.((((((...)))))).)))))))....))..)). ( -22.00) >DroPer_CAF1 16715 99 + 1 ---UAACAG-----C---CGCUUCUUGCAGGCGCGGGCAGAGUCAGCUCCUUUGCCUGCCCGAGCUGUUUGCCCGUGUGGACCUUGUCCAUACGGGAAACUACCGCAUCG ---.(((((-----(---(((.....)).((.((((((((((.......))))))))))))).))))))..(((((((((((...))))))))))).............. ( -47.90) >consensus ___UAACAG__UCGC___UGCUUCCUACAGACGCCGGCAGAGCCAGUUGCUUUGCCUGCCCGAACUGUUCGCCCGUGUGGACAUUGUCCAUGCGGGAAACUAUCGCAUCG .............................((.((.(((((((.......))))))).))............(((.(((((((...))))))).)))......))...... (-16.92 = -16.90 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:58 2006