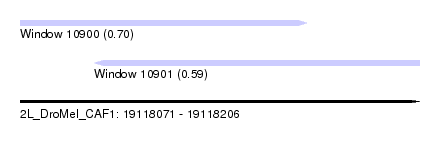
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,118,071 – 19,118,206 |
| Length | 135 |
| Max. P | 0.701244 |

| Location | 19,118,071 – 19,118,168 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 60.63 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -6.89 |
| Energy contribution | -7.53 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19118071 97 + 22407834 ACCAAUUCAGCAGGACGCGCACACGAACGACUCAAAC-GGAAACAACAAAAGUUGAGCCGCAUUCGGAGCGCAC----ACUGCGACCGGAU-UUCGAA-CCCGA .........((((...((((.(.((((((.(((....-(....)(((....)))))).))..))))).))))..----.))))...(((..-......-.))). ( -26.40) >DroPse_CAF1 10888 94 + 1 ACCAAUUC-GCGGGGUGGGC--CUAAACAGCUCACAG-GGAGACAACGAAAGUUGAGCCACAUU-GAGGCACAC----GCUGCGACAAUCGAUCCAAG-CAUAA ......((-((((.(((.((--((.....((((....-(....)(((....)))))))......-.)))).)))----.)))))).............-..... ( -32.70) >DroYak_CAF1 5229 97 + 1 ACCAAAUCAGCAGUACGCGCACACGAAGGACUCAAAC-GGAAACAACAAAAGUUGAGCCGCAUUCGGAGCGCAC----ACUGCGACCGGAU-UUCGAU-UCCGA .........(((((..((((.(.((((((.(((....-(....)(((....))))))))...))))).))))..----)))))...((((.-......-)))). ( -30.10) >DroMoj_CAF1 7238 78 + 1 ---UGCUC----AGAUGCUCGCACAAAUUGUUCAGUU-GGCAACAACAAAAGUUGAACAAUAUA---UAGGAAC----AGG--------UA-AUCAAAGCGC-- ---.(((.----.((((((..(....(((((((((((-(....))))......))))))))...---...)...----.))--------).-)))..)))..-- ( -16.50) >DroAna_CAF1 5259 101 + 1 ACCAAUUCAGCUGGAAGUGCACCCGAACGACCCAGAGGGGGAACAACAAAAGUUGAGCCGCAACCCGGGUGCACUUACUUUGUGACCAAGG-UUCGA--CGUGG .(((..((......(((((((((((.....(((...)))((..((((....))))..))......)))))))))))((((((....)))))-)..))--..))) ( -36.70) >DroPer_CAF1 10544 94 + 1 ACCAAUUC-GCGGGGUGGGC--CUAAACAGCUCACAG-GGAGACAACGAAAGUUGAGCCACAUU-GAGGCACAC----GCUGCGACAAUCGAUCCAAG-CAUAA ......((-((((.(((.((--((.....((((....-(....)(((....)))))))......-.)))).)))----.)))))).............-..... ( -32.70) >consensus ACCAAUUC_GCAGGAUGCGCACACAAACGACUCAAAG_GGAAACAACAAAAGUUGAGCCACAUU_GGGGCGCAC____ACUGCGACCAGAG_UUCAAA_CACGA .........((((..............................((((....)))).(((........))).........))))..................... ( -6.89 = -7.53 + 0.64)
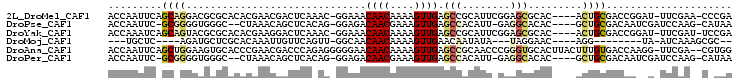
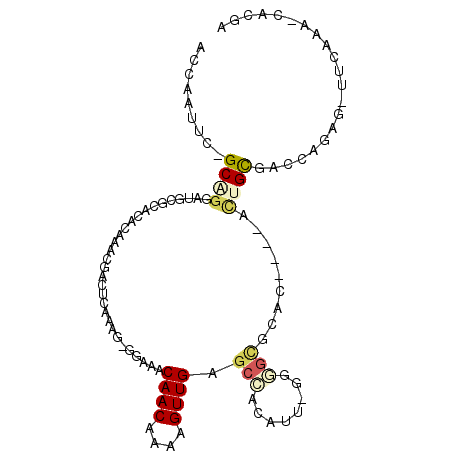
| Location | 19,118,096 – 19,118,206 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -21.17 |
| Energy contribution | -18.63 |
| Covariance contribution | -2.54 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19118096 110 - 22407834 UUUGUCUCAUGUAAUCGGUUUUUCCAUUAUCUUCCCGAUCGGGUUCGAAAUCCGGUCGCAGU----GUGCGCUCCGAAUGCGGCUCAACUUUUGUUGUUUCC-GUUUGAGUCGUU ...(.((((.(((.((((......(((...((...(((((((((.....))))))))).)).----)))....)))).)))((..((((....))))...))-...)))).)... ( -30.00) >DroYak_CAF1 5254 110 - 1 UUUGUCUCAUGUAAUCGGUUUUUCCAUUAUCUUCCCGAUCGGAAUCGAAAUCCGGUCGCAGU----GUGCGCUCCGAAUGCGGCUCAACUUUUGUUGUUUCC-GUUUGAGUCCUU ...(.((((.(((.((((......(((...((...((((((((.......)))))))).)).----)))....)))).)))((..((((....))))...))-...)))).)... ( -28.30) >DroAna_CAF1 5284 114 - 1 UUUGUCUCAUAUAAGUGGUUUUUCCAUUUUCAUACCGACCACG-UCGAACCUUGGUCACAAAGUAAGUGCACCCGGGUUGCGGCUCAACUUUUGUUGUUCCCCCUCUGGGUCGUU ............((((((.....))))))......(((((..(-((((((((.((((((.......))).))).))))).)))).((((....))))...........))))).. ( -32.50) >consensus UUUGUCUCAUGUAAUCGGUUUUUCCAUUAUCUUCCCGAUCGGG_UCGAAAUCCGGUCGCAGU____GUGCGCUCCGAAUGCGGCUCAACUUUUGUUGUUUCC_GUUUGAGUCGUU .((((...........((.....))...........(((((((.......))))))))))).........((((.((....((..((((....))))..))...)).)))).... (-21.17 = -18.63 + -2.54)

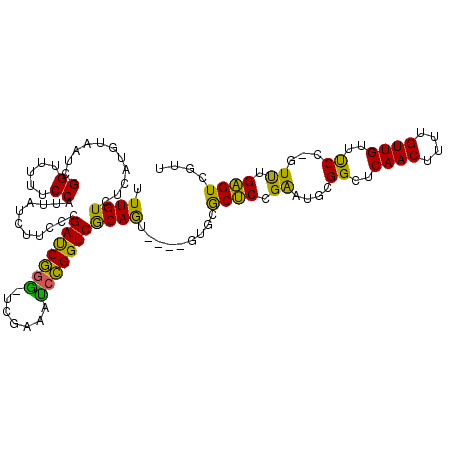
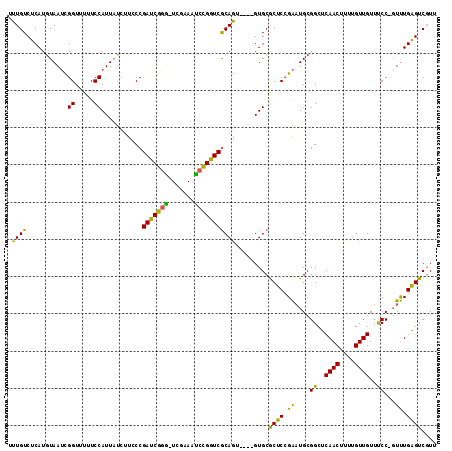
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:57 2006