| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,114,786 – 19,114,900 |
| Length | 114 |
| Max. P | 0.851557 |

| Location | 19,114,786 – 19,114,900 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -14.81 |
| Energy contribution | -16.12 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
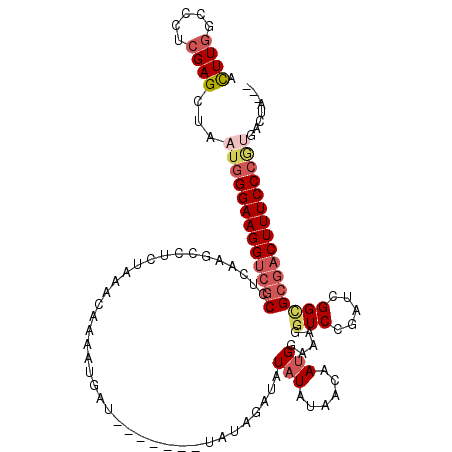
>2L_DroMel_CAF1 19114786 114 + 22407834 ACUUGGCCCUCGAGCUAAUGGGAAGGUCGCCUCAACCCCCUAAACAAAAUGAUAUAUAUAUAUAGAUAUAUAUAGCAAUGGAAAUGUCCGAUCGGUGCGACUUUCCCGUGACUA--- ...((((......))))(((((((((((((....(((............((.(((((((((....))))))))).)).((((....))))...)))))))))))))))).....--- ( -35.40) >DroSec_CAF1 2063 112 + 1 ACUUGGCCCUCGAGCUAAUGGGAAGGUCGCCUCAAACCUCUAAACAAAGUGAUAUAU--AUAUAGAUAUAUAUAGAUAUGGAAAGGUCCGAUCGGCGCGACUUUCCCGUGACUA--- ...((((......))))(((((((((((((.((..................((((((--((....)))))))).(((.((((....))))))))).))))))))))))).....--- ( -35.30) >DroSim_CAF1 2350 112 + 1 ACUUGGCCCUCGAGCUAAUGGGAAGGUCGCCUCAAACCUCUAAACAAAGUGAUAUAU--AUAUAGAUAUAUAUAGCAAUGAAAAGGUCCGAUCGGCGCGACUUUCCCGUGACUA--- ...((((......))))(((((((((((((.....((((.....((..((.((((((--((....)))))))).))..))...))))((....)).))))))))))))).....--- ( -33.30) >DroEre_CAF1 2004 101 + 1 ACUUGGCCCUCGAGUU---GGGAAGGUCGCCGCAAGUCUCUAAACAAAAG----------UACAAGUAUAUUUAACAAUGGAAAGGUCCGAUCGGCGCGACUUUCCCGUGACUA--- (((((.....))))).---(((((((((((.((..(((.........(((----------((......)))))......(((....))))))..))))))))))))).......--- ( -30.50) >DroYak_CAF1 2134 102 + 1 ACUUGGCCCUCGAGUUGAUGGGAAGGUCGCCUUACGUCUCUAAAUAAAAG----------UAUA--UAUAUUUAACAAUGGAAAGGUCCGAUCGGCGCCACUUUCCCGUGACUA--- ............((((.((((((((((((((.........((((((....----------....--..))))))....((((....))))...))))..)))))))))))))).--- ( -25.80) >DroPer_CAF1 7548 93 + 1 GUUUAGCCUUAGAGA-ACUAGGAAGGCCGCUGAGAGC-------------GAG--------CUG--GAUAUGCAACAAAGCAAAGGUCGGAUCGGAGAGACUUUCCCAUGACUAUAU ((((((((((((...-.))))).....(((.....))-------------).)--------)))--))).(((......)))..(((((....(((((...)))))..))))).... ( -22.20) >consensus ACUUGGCCCUCGAGCUAAUGGGAAGGUCGCCUCAAGCCUCUAAACAAAAUGAU_______UAUAGAUAUAUAUAACAAUGGAAAGGUCCGAUCGGCGCGACUUUCCCGUGACUA___ .((((.....))))...(((((((((((((......................................(((......))).....(((.....))))))))))))))))........ (-14.81 = -16.12 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:55 2006