| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,085,322 – 19,085,419 |
| Length | 97 |
| Max. P | 0.687055 |

| Location | 19,085,322 – 19,085,419 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.48 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
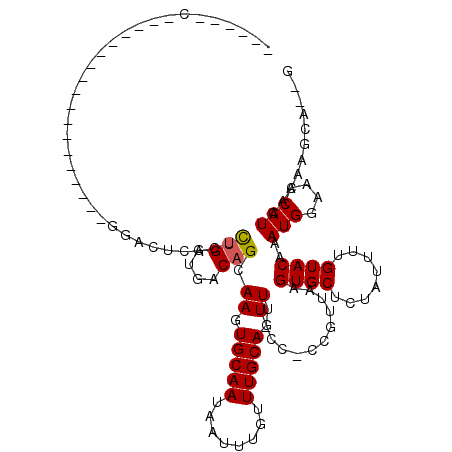
>2L_DroMel_CAF1 19085322 97 + 22407834 CAGACGC------------------AGAGUCAGUCUGGUGACAGCAAGUGCAAUAAUUUGUUUGCAGUUUUCC-CCGUUAAGUGCUCUAUUUUGUACAAAUGGAAACAUCACCAG----G ..(((..------------------...)))...(((((((..((((..((((....)))))))).((((...-(((((..((((........)))).))))))))).)))))))----. ( -28.00) >DroVir_CAF1 12116 93 + 1 ------U-------------------GACUCGCUCUGGUGACAGCAAGUGCAAUAAUUUGUUUGCAGUUU-CC-CCGUUAAGUGCUCUAUUUUGUACAAAUGGAAACAUCAGAGCCAGCG ------.-------------------(.((.(((((((((...((((..((((....)))))))).....-..-(((((..((((........)))).)))))...))))))))).))). ( -29.20) >DroGri_CAF1 12478 94 + 1 ------A------------------GGACUCGCUUUGGUGACAGCAAGUGCAAUAAUUUGUUUGCAGUUU-CA-ACGUUAAGUGCUCUAUUUUGUACAAAUGGAAACAUCAGAGGCAAAG ------.------------------..(((.(((..(....)))).)))((((........)))).((((-(.-.......((((........))))..(((....)))..))))).... ( -21.50) >DroWil_CAF1 13678 83 + 1 -------------------------------ACUCUGGUGACAGCAAGUGCAAUAAUUUGUUUGCAGUU-UCC-CCGUUAAGUGCUCUAUUUUGUACAAAUGGAAACAUCACCAG----C -------------------------------...(((((((..((((..((((....)))))))).(((-(((-.......((((........))))....)))))).)))))))----. ( -25.50) >DroMoj_CAF1 14273 95 + 1 ------C------------------GGACUCGCUUUGGUGACAGCAAGUGCAAUAAUUUGUUUGCAGUUU-CCCCCGUUAAGUGCUCUAUUUUGUACAAAUGGAAACAUCACAGUCAGAG ------.------------------...((.(((.(((((...((((..((((....)))))))).....-...(((((..((((........)))).)))))...))))).))).)).. ( -21.80) >DroAna_CAF1 8522 118 + 1 CACACACACAGCAACAAAUACAGUCGGAGUCAGUCCGGUGACAGCAAGUGCAAUAAUUUGUUUGCAGUUUUCC-CCGUUAAGUGCUCUAUUUUGUACAAAUGGAAACAUCAGAAGCUU-C ..........(((((((((.((.(((((.....)))))))...((....))....))))).))))((((((..-.......((((........))))..(((....)))..)))))).-. ( -24.10) >consensus ______C__________________GGACUCACUCUGGUGACAGCAAGUGCAAUAAUUUGUUUGCAGUUU_CC_CCGUUAAGUGCUCUAUUUUGUACAAAUGGAAACAUCACAAGCA__G ....................................((((...((((..((((....)))))))).........(((((..((((........)))).)))))...)))).......... (-15.32 = -15.48 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:51 2006