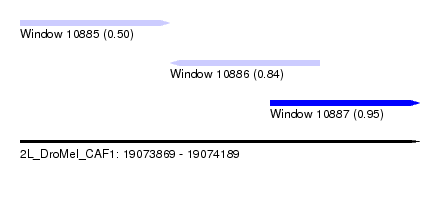
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,073,869 – 19,074,189 |
| Length | 320 |
| Max. P | 0.954299 |

| Location | 19,073,869 – 19,073,989 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -18.30 |
| Energy contribution | -17.54 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19073869 120 + 22407834 AGUAGUUUCACUGCAACCACCUCACCACGGCUGUUUCCCAAAAUCCUGAUGAUUCCACUGCUCAUGCGCAAAUCUACCUUAUUCUCCUCCAGGAGCUUCAGUAUCCGCUGUCCUAUCAAC .((((.....))))............(((((((.....)).....((((.((((....(((......))))))).........((((....))))..)))).....)))))......... ( -17.70) >DroSec_CAF1 6782 120 + 1 AGGAGCUCCACUGCAGCCACCUCACAACGGCUGUUUCCCAAAAUCCUGAUGAUUCCACUGCUCAUGCGCAAAUCUACCUUAUUCUCCUCCAGGAGCUUCAGUAUCCGCUGUCCUAUCAAC (((((((((...((((((..........))))))................((((....(((......))))))).................)))))..((((....))))))))...... ( -26.60) >DroSim_CAF1 6790 120 + 1 AGGAGCUCCACUGCAGCCACCUCCCCACGGCUGUUUCCCAAAAUCCUGAUGAUUCCACUGCUCAUGCGCAAAUCUACCUUAUUCUCCUCCAGGAGCUUCAGUAUCCGCUGUCCUAUCAAC (((((((((...((((((..........))))))................((((....(((......))))))).................)))))..((((....))))))))...... ( -26.60) >DroEre_CAF1 6146 120 + 1 AGGAGUUCCACUGCAGUCACUUCCCCACGAUUGUUUCCCAGGAUCCGGAUGAUUCCACUGCUCAUGCGCACAUCUACCUCAUGCUCCUCCAGGAGUUUCAGUAUCCGCUGUCCCACCAAC .((.........((((((..........))))))......((((.((((((.......(((......))).......((...(((((....)))))...))))))))..))))..))... ( -25.50) >DroYak_CAF1 6390 120 + 1 AGCAGCUUCACUGCAGUCACCUCCCCACGAUUGUUUCCCAGGAUCCGCGUGAUUCCACUGCUCACGCGCACUUCUACCUUAUUCUCCUCCAGGAGUUCCUGUAUCCGCUGUCCUAUCGAC .(((((......((((((..........))))))....(((((..(((((((.........)))))))...............((((....)))).))))).....)))))......... ( -31.30) >consensus AGGAGCUCCACUGCAGCCACCUCCCCACGGCUGUUUCCCAAAAUCCUGAUGAUUCCACUGCUCAUGCGCAAAUCUACCUUAUUCUCCUCCAGGAGCUUCAGUAUCCGCUGUCCUAUCAAC .((((((((...((((((..........))))))........................(((......))).....................))))))))..................... (-18.30 = -17.54 + -0.76)



| Location | 19,073,989 – 19,074,109 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -46.82 |
| Consensus MFE | -38.42 |
| Energy contribution | -37.74 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19073989 120 - 22407834 AGUGGUGUGCCUGGGUUCCAGUUUCAUGGCUGUGGAGGCUACGGCCAAUCUGGUUUCUAGAGCGCGCAGCGUUACGCUGGUAGCCCGGCAAAAUGUGCCUUUCAAAAGCACAUUGGGCGA ....(((((((((((..((((.....(((((((((...)))))))))..))))..))))).))))))..((((..(((((....)))))..(((((((.........))))))).)))). ( -52.00) >DroSec_CAF1 6902 120 - 1 AGUGGUGUGCCUGGGUUCCAGUUUCAUGGCUGUGGAGGCUUCGGCCAAUCUCGUUUCCAGAGCGGGCAGCGUUACCCUGGUAGCUCGACAAAAUGUGCCUUUCAAAAGCACAUUGGGUGA .((.((.((((((((((((((((....))))).)))(((....))).........))))).))).)).))(((((....)))))...((..(((((((.........)))))))..)).. ( -42.80) >DroSim_CAF1 6910 120 - 1 AGUGGUGUGCCUGGGUUCCAGUUUCAUGGCUGUGGAGGCUUCGGCCAAUCUCGUUUCCAGAGCGGGCAGCGUUACCCUGGUAGCCCGUCAAAAUGUGCCCUUCAAAAGCACAUUGGGUGA .((.((.((((((((((((((((....))))).)))(((....))).........))))).))).)).)).((((((.((....)).....(((((((.........))))))))))))) ( -42.90) >DroEre_CAF1 6266 120 - 1 AGUGGUCUGCCUGGGUUCCAGUUUCAUGGCUGUGGAAGCUUCCGCCAAUCUCGUUUCCAGAGCGGGCAGCGUUACCCUGGUAGCCCGACAAAAUGUGCCAUUCCAAAGCACAUUGGGUGA ..(((.(((((.((((..(.(((...(.(((.(((((((.............))))))).))).)..))))..)))).))))).)))((..(((((((.........)))))))..)).. ( -43.72) >DroYak_CAF1 6510 120 - 1 CGUGGUGUGCCUGGGCUCCAGUUUCAUGGCUGUGGAGGCUUCCGCCAAUCUCGUUGCCAGAGCGGGCCGCGUUACCCUGGUAGCGCGACAAAAUGUGCCUUUUAAAGGCACAUUGGGUGA ...(((.(((((((.(..(((((....)))))..).(((....)))..........)))).))).)))(((((((....))))))).((..(((((((((.....)))))))))..)).. ( -52.70) >consensus AGUGGUGUGCCUGGGUUCCAGUUUCAUGGCUGUGGAGGCUUCGGCCAAUCUCGUUUCCAGAGCGGGCAGCGUUACCCUGGUAGCCCGACAAAAUGUGCCUUUCAAAAGCACAUUGGGUGA ........((((((((((((........(((.(((((((.............))))))).)))(((........)))))).))))).....(((((((.........))))))))))).. (-38.42 = -37.74 + -0.68)

| Location | 19,074,069 – 19,074,189 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -27.90 |
| Energy contribution | -29.10 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19074069 120 + 22407834 AGCCUCCACAGCCAUGAAACUGGAACCCAGGCACACCACUUGGGUUGAUUUGUCCACCAUUUUGAAUAUGCUCCUGGCAUCGCCAAUAUUCCGAAUGACUUUCACAUUUUUCAAGUGCAC .((((......(((......))).....))))....((((((((..(((.((.....(((((.((((((.....((((...)))))))))).))))).....)).))).))))))))... ( -27.90) >DroSec_CAF1 6982 120 + 1 AGCCUCCACAGCCAUGAAACUGGAACCCAGGCACACCACUUUGGUUGAUUUGUCCACCAUUUGGAAUAUGCUCCUGGCAUCGCCAACGUCCCGAAUGGUUUUCACAUUUUUCAAGUGCAU .((((......(((......))).....))))....(((((.((..(((.((...(((((((((...(((....((((...)))).))).)))))))))...)).)))..)))))))... ( -30.20) >DroSim_CAF1 6990 120 + 1 AGCCUCCACAGCCAUGAAACUGGAACCCAGGCACACCACUUGGGUUGAUUUGUCCACCAUUUGGAAUAUGCUCCUGGCAUCGCCAAUGUCCCGAAUGGUUUUCACAUUUUUCAAGUGCAC .((((......(((......))).....))))....((((((((..(((.((...(((((((((.((((.....((((...)))))))).)))))))))...)).))).))))))))... ( -34.60) >DroEre_CAF1 6346 120 + 1 AGCUUCCACAGCCAUGAAACUGGAACCCAGGCAGACCACUUGCGUUGAUUUGCCCACCAUUUGGAAUAUGCUCCUGGCGUCGCCAAUAUCCCGAAUGGUUUUCACAUUUUUCAAGUGCAC ..........(((.(((((.(((((....(((((((.((....)).).)))))).(((((((((.((((.....((((...)))))))).))))))))).))).))..))))).).)).. ( -29.80) >DroYak_CAF1 6590 120 + 1 AGCCUCCACAGCCAUGAAACUGGAGCCCAGGCACACCACGUGCGUUGAUUUGUCCACCAUUUGGAAUAUGCUCCUGGCAUCGCCAAUAUCCCGAAUGGUUUUCACAUUCUUCAAGUGCAC .((((......(((......))).....)))).......(((((((((..(((..(((((((((.((((.....((((...)))))))).)))))))))....)))....)))).))))) ( -33.70) >consensus AGCCUCCACAGCCAUGAAACUGGAACCCAGGCACACCACUUGGGUUGAUUUGUCCACCAUUUGGAAUAUGCUCCUGGCAUCGCCAAUAUCCCGAAUGGUUUUCACAUUUUUCAAGUGCAC .((((......(((......))).....))))....((((((((..(((.((...(((((((((.((((.....((((...)))))))).)))))))))...)).))).))))))))... (-27.90 = -29.10 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:44 2006