
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,073,052 – 19,073,159 |
| Length | 107 |
| Max. P | 0.998690 |

| Location | 19,073,052 – 19,073,159 |
|---|---|
| Length | 107 |
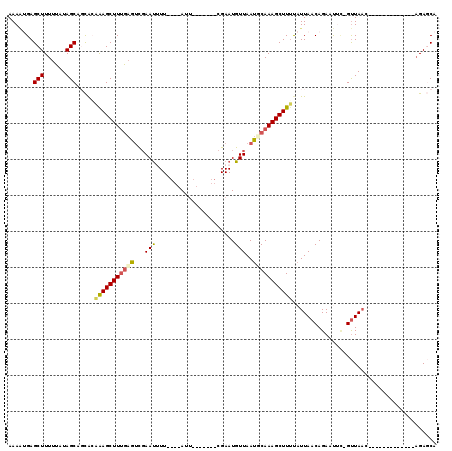
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -10.29 |
| Energy contribution | -10.41 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.42 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
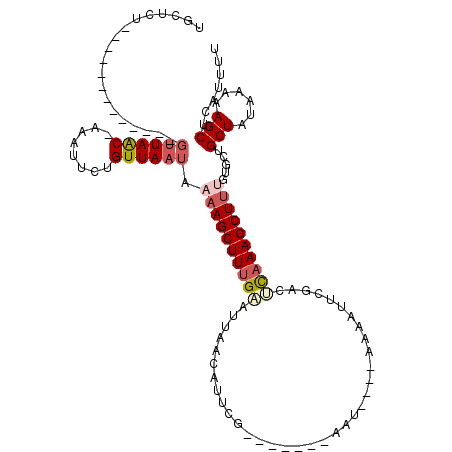
>2L_DroMel_CAF1 19073052 107 + 22407834 AAAAUGAGCUUUUUAUAGCAGCACAAAGCUUUGAGUCGAAUUUU----AUU-------CGAAUGUUAAUACAAAGCUUUUAUUAACAGAAUUA-GUUAACAGAAUUAGUUAACAGAGCA .......(((......))).((..(((((((((..((((((...----)))-------)))((....)).)))))))))..............-((((((.......))))))...)). ( -21.50) >DroSec_CAF1 5993 104 + 1 AAAAUGAGCUUUUUAUAGCAGCAGAAAGCUUUGAGUUGAAUUUUCGACAUUCGAAAUUCGAAUGUUAAUUCAAAGCUUUUAUUAACAGAAUUC-GUUAAC--------------GAGCA .......(((......))).((.((((((((((((((((....))((((((((.....))))))))))))))))))))))...........((-(....)--------------)))). ( -31.40) >DroSim_CAF1 6007 98 + 1 AAAAUGAGCUUUUUAUAGCAGCAGAAAGCUUUGAGUCGAAUUUUCGACAUU-------CGAAUGUUAAUUCAAAGCUUUUAUUAACAGAAUUC-GUUAAC-------------AGAGCA .......(((......))).((.(((((((((((((..(...((((.....-------))))..)..))))))))))))).(((((.......-))))).-------------...)). ( -26.90) >DroEre_CAF1 5347 94 + 1 AAAGAGAGCUUUUUAUAGCACUACUAAGCUUUGAGUC-AAUGUU----AUA-------CGAAUGUUAAUGCAAAGCUUGCAUUAACAGAACAUAGCUAAC-------------AGACCA ....(((((((..((......))..)))))))(.(((-...(((----((.-------....((((((((((.....))))))))))....)))))....-------------.)))). ( -21.90) >DroYak_CAF1 5627 94 + 1 AAACGGAGCUUUUUAUAGCACUGCUGAGCUUAACGCCGAACGUU----AUU-------CGAAUGUUAACGCAAAGCUUUUAUUAACAGAACAU-GUUAUA-------------AGACCA ....((.(((......)))...(((..((((((((.((((....----.))-------))..)))))).))..)))((((((.((((.....)-))))))-------------))))). ( -19.40) >consensus AAAAUGAGCUUUUUAUAGCAGCACAAAGCUUUGAGUCGAAUUUU____AUU_______CGAAUGUUAAUGCAAAGCUUUUAUUAACAGAAUUC_GUUAAC_____________AGAGCA .......(((......))).....(((((((((((...(((......................)))..)))))))))))........................................ (-10.29 = -10.41 + 0.12)


| Location | 19,073,052 – 19,073,159 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -8.05 |
| Energy contribution | -9.17 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19073052 107 - 22407834 UGCUCUGUUAACUAAUUCUGUUAAC-UAAUUCUGUUAAUAAAAGCUUUGUAUUAACAUUCG-------AAU----AAAAUUCGACUCAAAGCUUUGUGCUGCUAUAAAAAGCUCAUUUU .((.(.((((((.......))))))-..............(((((((((.........(((-------(((----...))))))..)))))))))).)).(((......)))....... ( -19.30) >DroSec_CAF1 5993 104 - 1 UGCUC--------------GUUAAC-GAAUUCUGUUAAUAAAAGCUUUGAAUUAACAUUCGAAUUUCGAAUGUCGAAAAUUCAACUCAAAGCUUUCUGCUGCUAUAAAAAGCUCAUUUU .((..--------------((((((-(.....))))))).((((((((((....(((((((.....))))))).((....))...))))))))))..)).(((......)))....... ( -24.50) >DroSim_CAF1 6007 98 - 1 UGCUCU-------------GUUAAC-GAAUUCUGUUAAUAAAAGCUUUGAAUUAACAUUCG-------AAUGUCGAAAAUUCGACUCAAAGCUUUCUGCUGCUAUAAAAAGCUCAUUUU .((..(-------------((((((-(.....))))))))((((((((((.....(....)-------...(((((....)))))))))))))))..)).(((......)))....... ( -21.60) >DroEre_CAF1 5347 94 - 1 UGGUCU-------------GUUAGCUAUGUUCUGUUAAUGCAAGCUUUGCAUUAACAUUCG-------UAU----AACAUU-GACUCAAAGCUUAGUAGUGCUAUAAAAAGCUCUCUUU .(((((-------------((((..((((...((((((((((.....))))))))))..))-------)))----))))..-))))...(((((..((......))..)))))...... ( -22.40) >DroYak_CAF1 5627 94 - 1 UGGUCU-------------UAUAAC-AUGUUCUGUUAAUAAAAGCUUUGCGUUAACAUUCG-------AAU----AACGUUCGGCGUUAAGCUCAGCAGUGCUAUAAAAAGCUCCGUUU .((.((-------------(.((((-(.....)))))......(((..((.(((((..(((-------(((----...)))))).)))))))..)))...........)))..)).... ( -17.00) >consensus UGCUCU_____________GUUAAC_AAAUUCUGUUAAUAAAAGCUUUGAAUUAACAUUCG_______AAU____AAAAUUCGACUCAAAGCUUUGUGCUGCUAUAAAAAGCUCAUUUU ...................((((((........)))))).((((((((((...................................)))))))))).....(((......)))....... ( -8.05 = -9.17 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:41 2006