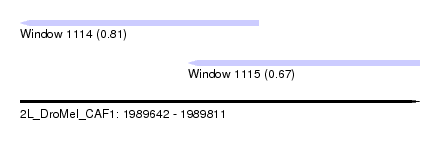
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,989,642 – 1,989,811 |
| Length | 169 |
| Max. P | 0.810019 |

| Location | 1,989,642 – 1,989,743 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -20.29 |
| Energy contribution | -19.85 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.810019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1989642 101 - 22407834 GGCUAUUCUUUGUUCAGAAGAACGGAGAGCAGAAAACGAGAAUGCGAAUGCGAAUACGAAUCCGAAUGCGAACUGU--AGACCGUAAUCGAAA------CCAAAACACU ((((.((((((((((....)))))))))).))............(((.((((..((((..((.......))..)))--)...)))).)))...------))........ ( -20.80) >DroSec_CAF1 7390 101 - 1 GGCUAUUCUUUGUUCGGAAGAACGGAGAGCAGAAAACGAGAAUGCGAAUGCGAAUACGAAUCCGAAUGCGAACUCU--AGACCGUAAUCGAAA------CCAAAACACU ((((.((((((((((....)))))))))).))....((....(((....)))....))....(((.((((......--....)))).)))...------))........ ( -22.70) >DroSim_CAF1 5262 101 - 1 GGCUAUUCUUUGUUCGGAAGAACGGAGAACAGAAAACGAGAAUGCGAAUGCGAAUAAGAAUCCGAAUGCGAACUCU--AGACCGUAAUCGAAA------UCAAAACACU ..((.((((((((((....)))))))))).)).......((...(((.((((....(((...((....))...)))--....)))).)))...------))........ ( -24.00) >DroYak_CAF1 8240 97 - 1 GGCUAUUCUUUGUUCGCAAGAACGGAGAACAGAAAACGAGAAUGCGA------------UUCCGAGUGCGAACUUUAUAAAGCGUAAUCGAAACCGAAACCAAAACACU ((((.((((((((((....)))))))))).))....(((..((((..------------....((((....))))......))))..)))...)).............. ( -26.90) >consensus GGCUAUUCUUUGUUCGGAAGAACGGAGAACAGAAAACGAGAAUGCGAAUGCGAAUACGAAUCCGAAUGCGAACUCU__AGACCGUAAUCGAAA______CCAAAACACU ((((.((((((((((....)))))))))).))..............................(((.((((............)))).))).........))........ (-20.29 = -19.85 + -0.44)

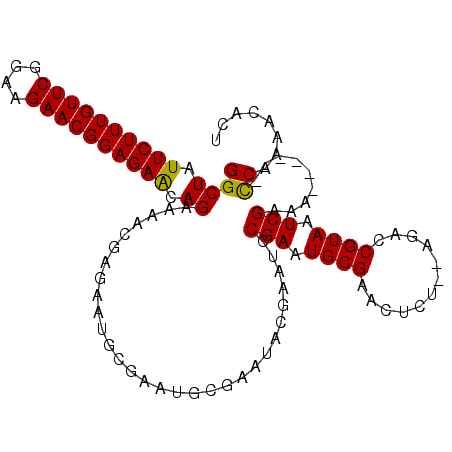
| Location | 1,989,713 – 1,989,811 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.07 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
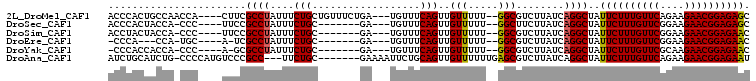
>2L_DroMel_CAF1 1989713 98 - 22407834 ACCCACUGCCAACCA----CUUCGCCUAUUUCUGCUGUUUCUGA---UGUUUCAGUUGUUUUU--GGCGUCUUAUCAGGCUAUUCUUUGUUCAGAAGAACGGAGAGC .......(((.....----...((((.......((((...(...---.)...)))).......--))))........)))..((((((((((....)))))))))). ( -25.03) >DroSec_CAF1 7461 90 - 1 ACCCACUACCA-CCC----UUCCGCCUAUUUCUGC-------GA---UGUUUCAGUUGUUUUU--GGCUUCUUAUCAGGCUAUUCUUUGUUCGGAAGAACGGAGAGC ...........-...----....(((.......((-------((---(......)))))....--)))..............((((((((((....)))))))))). ( -20.50) >DroSim_CAF1 5333 90 - 1 ACCUACUACCA-CCC----UUCCGCCUAUUUCUGC-------GA---UGUUUCAGUUGUUUUU--GGCGUCUUAUCAGGCUAUUCUUUGUUCGGAAGAACGGAGAAC ...........-...----....(((.......((-------((---(......)))))....--)))((((....))))..((((((((((....)))))))))). ( -23.40) >DroEre_CAF1 4292 85 - 1 -CCCA---CCA-UGC----A-UCGCCUAUUUCUGC-------GA---UGUUUCAGUUGUUUUU--GGCGUCUUAUCAGGCUAUUCUUUGUUCGGAAGAACGGAGAAC -....---(((-.((----(-((((........))-------))---)))............)--)).((((....))))..((((((((((....)))))))))). ( -25.72) >DroYak_CAF1 8307 88 - 1 -CCCACCACCA-CCC----A-GCGCCUAUUUCUGC-------GA---UGUUUCAGUUGUUUUU--GGCGUCUUAUCAGGCUAUUCUUUGUUCGCAAGAACGGAGAAC -....((....-...----.-(((((.......((-------((---(......)))))....--))))).......))...((((((((((....)))))))))). ( -24.26) >DroAna_CAF1 4816 96 - 1 AUCUGCAUCUG-CCCCAUGUCCCGCC---UUCUGC-------GAAAAUUCUGCAGUUGUUUUUUGAGCGUCUUAUCAGGCUAUUCUUUGUUCAGAAGAACGGAGAAU ..........(-((..(((...((((---..((((-------(.......))))).........).)))...)))..))).(((((((((((....))))))))))) ( -21.60) >consensus ACCCACUACCA_CCC____UUCCGCCUAUUUCUGC_______GA___UGUUUCAGUUGUUUUU__GGCGUCUUAUCAGGCUAUUCUUUGUUCGGAAGAACGGAGAAC .......................((((....(((..................)))..(((.....)))........))))..((((((((((....)))))))))). (-16.26 = -16.07 + -0.19)
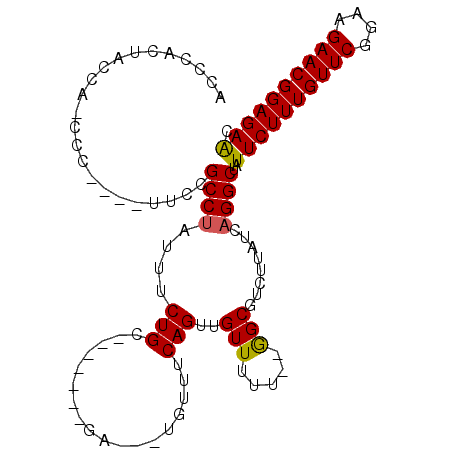
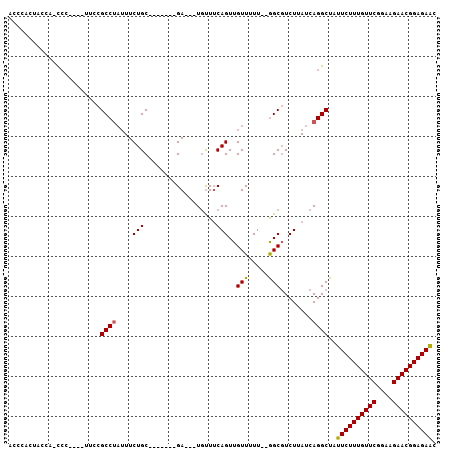
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:47 2006