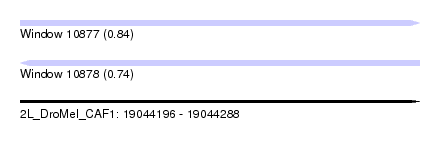
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,044,196 – 19,044,288 |
| Length | 92 |
| Max. P | 0.841176 |

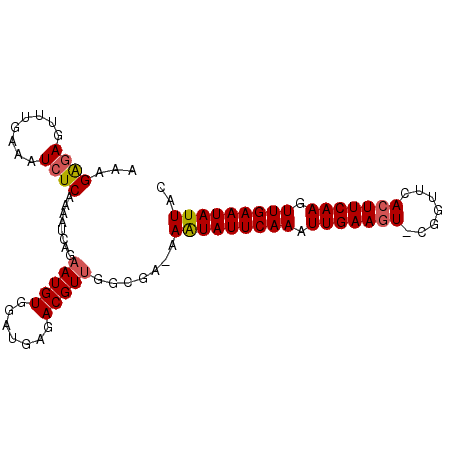
| Location | 19,044,196 – 19,044,288 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 87.77 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.71 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19044196 92 + 22407834 GUAAUAUUCAACUUGAACUGAACCG-ACUUCAAAUUGAAUACUU-UCGCCAACGUCUCAUCCACAUUCUGAUUUUGAGAUUUCAAACUCUCUUU ....(((((((.(((((........-..))))).)))))))...-........((((((...............)))))).............. ( -11.76) >DroSec_CAF1 12415 92 + 1 GUAAUAUUCAACUUGAAGUGAACCG-ACUUCAAAUUGAAUAUUU-UGGCCAACGUCUCACCCACAUUCUGAUUUUGAGAUUUCAAACUCUCUUU ..(((((((((.(((((((......-))))))).))))))))).-........((((((...............)))))).............. ( -18.26) >DroSim_CAF1 10757 92 + 1 GUAAUAUUCAACUUGAAGUGAACCG-ACUUCAAAUUGAAUAUUU-UGGCCAACGUCUCACCCACAUUCUGAUUUUGAGAUUUCAAACUCUCUUU ..(((((((((.(((((((......-))))))).))))))))).-........((((((...............)))))).............. ( -18.26) >DroEre_CAF1 5943 92 + 1 GUAAUAUUCAACUUGAAGUGAACAG-ACUUCAAUUUGAAUAUUU-UCGCCAACGUCCCAUCCACAUUCUGAUUUUGAGAUUUUGGACUCUCUUU ..(((((((((.(((((((......-))))))).))))))))).-...((((.(((.((...............)).))).))))......... ( -20.36) >DroYak_CAF1 5863 92 + 1 GUAAUAUUCAACUUGAAGUGAACAG-ACUUCAAUUUGAAUAUUU-UCGCCAACGUCUCAUCCACAUUCUGAUUUUGAGAUUUCGAACUCUCUUU ..(((((((((.(((((((......-))))))).)))))))))(-(((.....((((((...............))))))..))))........ ( -20.76) >DroAna_CAF1 12958 90 + 1 GUAAUAUUCAACUUGAAGUCAAAUGUACUUCAAUUUGAAUAUUUUUUACUGACGUUUU---GACAUUCUAACUUUGAGAUU-CAAAUUACCCUU ..(((((((((.((((((((....).))))))).)))))))))............(((---((..(((.......)))..)-))))........ ( -16.80) >consensus GUAAUAUUCAACUUGAAGUGAACCG_ACUUCAAAUUGAAUAUUU_UCGCCAACGUCUCAUCCACAUUCUGAUUUUGAGAUUUCAAACUCUCUUU ..(((((((((.(((((((.......))))))).)))))))))..........((((((...............)))))).............. (-14.32 = -14.71 + 0.39)

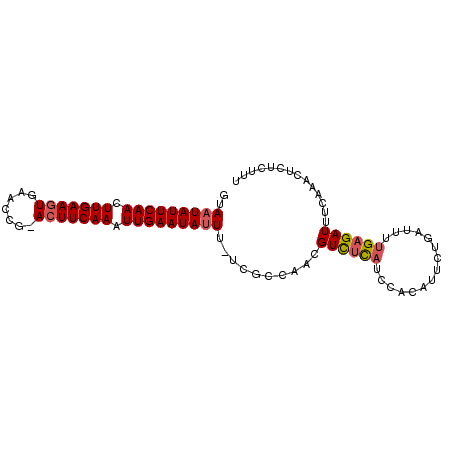
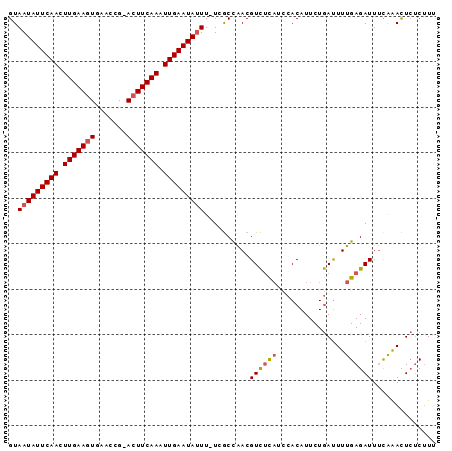
| Location | 19,044,196 – 19,044,288 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 87.77 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19044196 92 - 22407834 AAAGAGAGUUUGAAAUCUCAAAAUCAGAAUGUGGAUGAGACGUUGGCGA-AAGUAUUCAAUUUGAAGU-CGGUUCAGUUCAAGUUGAAUAUUAC ...((((........))))..........(((.((((...)))).))).-.(((((((((((((((.(-......).))))))))))))))).. ( -22.20) >DroSec_CAF1 12415 92 - 1 AAAGAGAGUUUGAAAUCUCAAAAUCAGAAUGUGGGUGAGACGUUGGCCA-AAAUAUUCAAUUUGAAGU-CGGUUCACUUCAAGUUGAAUAUUAC .......(.(..(..(((((...(((.....))).)))))..)..).).-.(((((((((((((((((-......))))))))))))))))).. ( -26.20) >DroSim_CAF1 10757 92 - 1 AAAGAGAGUUUGAAAUCUCAAAAUCAGAAUGUGGGUGAGACGUUGGCCA-AAAUAUUCAAUUUGAAGU-CGGUUCACUUCAAGUUGAAUAUUAC .......(.(..(..(((((...(((.....))).)))))..)..).).-.(((((((((((((((((-......))))))))))))))))).. ( -26.20) >DroEre_CAF1 5943 92 - 1 AAAGAGAGUCCAAAAUCUCAAAAUCAGAAUGUGGAUGGGACGUUGGCGA-AAAUAUUCAAAUUGAAGU-CUGUUCACUUCAAGUUGAAUAUUAC .......(.((((..(((((...(((.....))).)))))..)))))..-.(((((((((.(((((((-......))))))).))))))))).. ( -23.50) >DroYak_CAF1 5863 92 - 1 AAAGAGAGUUCGAAAUCUCAAAAUCAGAAUGUGGAUGAGACGUUGGCGA-AAAUAUUCAAAUUGAAGU-CUGUUCACUUCAAGUUGAAUAUUAC ...((((........))))..........(((.((((...)))).))).-.(((((((((.(((((((-......))))))).))))))))).. ( -22.40) >DroAna_CAF1 12958 90 - 1 AAGGGUAAUUUG-AAUCUCAAAGUUAGAAUGUC---AAAACGUCAGUAAAAAAUAUUCAAAUUGAAGUACAUUUGACUUCAAGUUGAAUAUUAC ........((((-(...((.......))...))---)))............(((((((((.(((((((.(....)))))))).))))))))).. ( -17.50) >consensus AAAGAGAGUUUGAAAUCUCAAAAUCAGAAUGUGGAUGAGACGUUGGCGA_AAAUAUUCAAAUUGAAGU_CGGUUCACUUCAAGUUGAAUAUUAC ...((((........))))........(((((.......))))).......(((((((((.(((((((.......))))))).))))))))).. (-17.48 = -17.70 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:36 2006