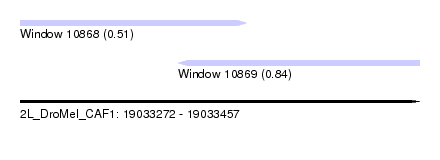
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,033,272 – 19,033,457 |
| Length | 185 |
| Max. P | 0.840334 |

| Location | 19,033,272 – 19,033,377 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19033272 105 + 22407834 UGGAGAGUUUAUUUUUGGACCUUGGUAUUCCUCCGACUGG-------GGUGUGUUGAAAAUAGACAUAAACAAAUUUUCAGAUGGGAGCCCAGCCAUCGGUGCUCCC--------AUUCC .((((.(((((((((..(((((..((.........))..)-------)))...)..))))))))).................(((((((((.......)).))))))--------))))) ( -35.00) >DroSim_CAF1 34938 105 + 1 UGGAGAGUUUAUUUUUGGACCUUGGUAUUCCUCCGACUGG-------GGUGUGUUGAAAAUAGACAUAAACAAAUUUUCAGAUGGGAGCCCAGCCAGCGGUGCUCCC--------AUUCC .((((.(((((((((..(((((..((.........))..)-------)))...)..))))))))).................(((((((((.......)).))))))--------))))) ( -35.00) >DroEre_CAF1 35456 113 + 1 UGGAGAGUUUAUUUUUGGACCUUGGUAUUCCUCCGACUGG-------GGUGUGUUGAAAAUAGACAUAAACAAAAUUUCAGAUGGCGGCCCUGCCAGCGGUGCUCCCAUUCUCCCAUUCC .((((((((((((((..(((((..((.........))..)-------)))...)..)))))))))................((((.(((.(((....))).))).)))))))))...... ( -34.90) >DroYak_CAF1 31604 112 + 1 UGGAGAGUUUAUUUUUGGACUUUGGUAUUCUUCCGACUGGGGUCUGGGGUGUGUUGAAAAUAGACAUAAACAAAAUUUCAGAUGGCAGCCCAGCCAGCGGUGCUCCC--------AUUCC .((((((((((....))))))...........(((.((((...(((((.(((.((((((................))))))...))).))))))))))))..)))).--------..... ( -31.79) >consensus UGGAGAGUUUAUUUUUGGACCUUGGUAUUCCUCCGACUGG_______GGUGUGUUGAAAAUAGACAUAAACAAAAUUUCAGAUGGCAGCCCAGCCAGCGGUGCUCCC________AUUCC .((((.(((((((((..((((((((.......))))...........)))...)..)))))))))..................((((((((.......)).)))))).........)))) (-23.60 = -23.73 + 0.12)

| Location | 19,033,345 – 19,033,457 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -44.39 |
| Consensus MFE | -39.47 |
| Energy contribution | -40.48 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19033345 112 - 22407834 CUGCUCUAUAAUUACCCCGUAUCACGGGUGUGGAUGUGGAAUCGUUCUCUAGAGGGUGACAUUUGAUGUUGGCCUGGGGAGGAAU--------GGGAGCACCGAUGGCUGGGCUCCCAUC ....((((((.((((((((.....)))).)))).))))))....((((((..(((.((((((...)))))).)))..))))))((--------((((((.(((.....))))))))))). ( -47.20) >DroSim_CAF1 35011 112 - 1 CUGCUCUAUAAUUACCCCGUAUCACGGGUGUGGAUGUGGAAUCGUUCUCUAGAGGGUGACAUUUGAUGUUGGCCUGGGGAGGAAU--------GGGAGCACCGCUGGCUGGGCUCCCAUC ....((((((.((((((((.....)))).)))).))))))....((((((..(((.((((((...)))))).)))..))))))((--------((((((.(((.....))))))))))). ( -46.90) >DroEre_CAF1 35529 120 - 1 CUGCUCUAUAAUUACCCCGUAUCACGGGUGUGGAUGUGGAAUCGUUCUGUAGAGGGUGACAUUUGAUGUUGGCCUGGGGAGGAAUGGGAGAAUGGGAGCACCGCUGGCAGGGCCGCCAUC ..((((((((.((((((((.....)))).)))).))))))...(((((((..((((((.(((((........(((....))).......)))))....)))).)).))))))).)).... ( -38.26) >DroYak_CAF1 31684 112 - 1 CUGCUCUAUAAUUACCCCGUAUCACGGGUGUGGAUGUGGAAGCGUUCUCUAGAGGGUGACAUUUGAUGUUGGCCUGGGGAGGAAU--------GGGAGCACCGCUGGCUGGGCUGCCAUC ..((((((((.((((((((.....)))).)))).))))).))).((((((..(((.((((((...)))))).)))..))))))((--------((.(((.(((.....)))))).)))). ( -45.20) >consensus CUGCUCUAUAAUUACCCCGUAUCACGGGUGUGGAUGUGGAAUCGUUCUCUAGAGGGUGACAUUUGAUGUUGGCCUGGGGAGGAAU________GGGAGCACCGCUGGCUGGGCUCCCAUC ....((((((.((((((((.....)))).)))).))))))....((((((..(((.((((((...)))))).)))..))))))..........((((((.((.......))))))))... (-39.47 = -40.48 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:28 2006