
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,030,981 – 19,031,075 |
| Length | 94 |
| Max. P | 0.818492 |

| Location | 19,030,981 – 19,031,075 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -12.74 |
| Energy contribution | -15.05 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
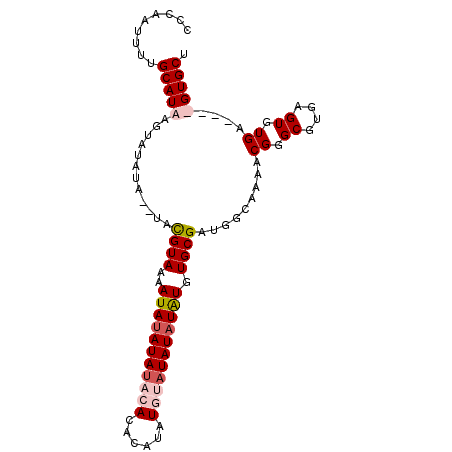
>2L_DroMel_CAF1 19030981 94 + 22407834 CUCAAUUUUGCAUAAGUAUAUA--UUUGUAAAAUAUAUACACAUACGUAU--AUAUAUGUGUGCGAUGGCAAAACGGGCGUGAGUGUGAGUGUGUGCU ((((.((((((....(((((((--((.....))))))))).(((.(((((--((....))))))))))))))))...((....)).))))........ ( -26.50) >DroSim_CAF1 32627 97 + 1 CCC-AUUUUGCAUAAGUAUAUAUGUUUGUAAAAUAUAUAUACACACAUAUGAAUAUAUAUGUGCGAUGGCAAAACGGGCGUGAGUGUGAGUGUGUGCU (((-.((((((....(((((((((((.....))))))))))).((((((((....)))))))).....)))))).)))....((..(......)..)) ( -27.30) >DroEre_CAF1 33227 90 + 1 CCCAAUUUUGCAUCAGUAUAU----ACGUAAAAUACAUAUAUACACAUAUUUAUAUAUAUGUGCGAUGUCAAAUCGGGCGUGAGUGUGA----GUGCU .........((....((((((----(.(((...))).)))))))....((((((((.(((((.((((.....)))).))))).))))))----)))). ( -23.90) >DroYak_CAF1 29180 91 + 1 CAAAAUUUUGCAUUAGUAUAUAUGUACGUAAAAUAUAUAUACAC---UGUGUAUAUAUAUGUGCGAUGUCAAAACGGGCGAUUGUGUGA----GUGCU .........(((((..((((..((((((((...(((((((((..---.))))))))))))))))).((((......))))..))))..)----)))). ( -21.30) >consensus CCCAAUUUUGCAUAAGUAUAUA__UACGUAAAAUAUAUAUACACACAUAUGUAUAUAUAUGUGCGAUGGCAAAACGGGCGUGAGUGUGA____GUGCU .........(((((..(((((.....((((..(((((((((((......))))))))))).))))(((((......)))))..)))))....))))). (-12.74 = -15.05 + 2.31)

| Location | 19,030,981 – 19,031,075 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -8.57 |
| Energy contribution | -9.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19030981 94 - 22407834 AGCACACACUCACACUCACGCCCGUUUUGCCAUCGCACACAUAUAU--AUACGUAUGUGUAUAUAUUUUACAAA--UAUAUACUUAUGCAAAAUUGAG ..............((((.....(((((((........((((((..--....))))))(((((((((.....))--)))))))....))))))))))) ( -17.60) >DroSim_CAF1 32627 97 - 1 AGCACACACUCACACUCACGCCCGUUUUGCCAUCGCACAUAUAUAUUCAUAUGUGUGUAUAUAUAUUUUACAAACAUAUAUACUUAUGCAAAAU-GGG ....................((((((((((....((((((((......))))))))((((((((.((.....)).))))))))....)))))))-))) ( -26.30) >DroEre_CAF1 33227 90 - 1 AGCAC----UCACACUCACGCCCGAUUUGACAUCGCACAUAUAUAUAAAUAUGUGUAUAUAUGUAUUUUACGU----AUAUACUGAUGCAAAAUUGGG .....----...........((((((((..((((((((((((......))))))))(((((((((...)))))----))))...))))..)))))))) ( -27.60) >DroYak_CAF1 29180 91 - 1 AGCAC----UCACACAAUCGCCCGUUUUGACAUCGCACAUAUAUAUACACA---GUGUAUAUAUAUUUUACGUACAUAUAUACUAAUGCAAAAUUUUG .....----..............((((((.(((.....((((((((((...---..)))))))))).....(((......)))..))))))))).... ( -11.80) >consensus AGCAC____UCACACUCACGCCCGUUUUGACAUCGCACAUAUAUAUACAUAUGUGUGUAUAUAUAUUUUACAAA__UAUAUACUUAUGCAAAAUUGGG .(((...............((.............))..((((((((((((....))))))))))))....................)))......... ( -8.57 = -9.07 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:24 2006