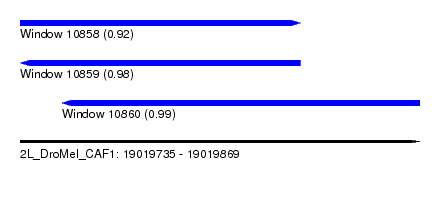
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,019,735 – 19,019,869 |
| Length | 134 |
| Max. P | 0.990605 |

| Location | 19,019,735 – 19,019,829 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19019735 94 + 22407834 --------------------------AAUAGAUAACGAUAUAUUUAUUUAUGCCAAUAAAACAGGUGAAAUUACCCAUAGUGCGGCGUAUUGUACGUGCUCUUACACCGCACAGUGUGGC --------------------------.((((((((........))))))))((((........((((....))))(((.((((((.(((..((....))...))).)))))).))))))) ( -26.00) >DroSec_CAF1 23761 117 + 1 GAAAUACAAA---UCAUCGUAUCUUAAGUAGUUACCGAUAUAUUUAUUUAUGCCAACAAAACAGGUGAAGUUGCCCAUAGUGCGGCGUAUUGUACGUGCUCUUACACCGCACAGUGUGGC ..........---((((((((.((.....)).))).(.((((......)))).).........)))))....((((((.((((((.(((..((....))...))).)))))).))).))) ( -27.90) >DroSim_CAF1 21368 120 + 1 GAAAUACAAAUAAUCAUCGUAUCUUAAGUAGUUAACAAUAUGUUUAUUUUAGCCAACAAAACAGGUGAAAUUGCCCAUAGUGCGGCGUAUUGUACGUGCUCUUACACCGCACAGUGUGGC .............(((((............(((((.((((....)))))))))..........)))))....((((((.((((((.(((..((....))...))).)))))).))).))) ( -27.65) >consensus GAAAUACAAA___UCAUCGUAUCUUAAGUAGUUAACGAUAUAUUUAUUUAUGCCAACAAAACAGGUGAAAUUGCCCAUAGUGCGGCGUAUUGUACGUGCUCUUACACCGCACAGUGUGGC ...................................................((((........(((......)))(((.((((((.(((..((....))...))).)))))).))))))) (-23.22 = -23.00 + -0.22)

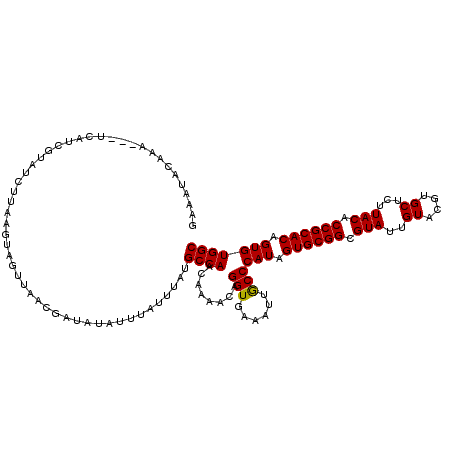
| Location | 19,019,735 – 19,019,829 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -24.49 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19019735 94 - 22407834 GCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGUAAUUUCACCUGUUUUAUUGGCAUAAAUAAAUAUAUCGUUAUCUAUU-------------------------- ((((....((((((((((.............)))))))))).((((((......)))))).....)))).........................-------------------------- ( -25.02) >DroSec_CAF1 23761 117 - 1 GCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGCAACUUCACCUGUUUUGUUGGCAUAAAUAAAUAUAUCGGUAACUACUUAAGAUACGAUGA---UUUGUAUUUC (((.((..((((((((((.............))))))))))..)))))......((((((((((((......)))))).)))..))).........((((((((...---.)))))))). ( -31.32) >DroSim_CAF1 21368 120 - 1 GCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGCAAUUUCACCUGUUUUGUUGGCUAAAAUAAACAUAUUGUUAACUACUUAAGAUACGAUGAUUAUUUGUAUUUC (((.((..((((((((((.............))))))))))..)))))..........((...((((((...............)))))).))...((((((((.......)))))))). ( -31.28) >consensus GCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGCAAUUUCACCUGUUUUGUUGGCAUAAAUAAAUAUAUCGUUAACUACUUAAGAUACGAUGA___UUUGUAUUUC (((.((..((((((((((.............))))))))))..)))))............((((((......))))))..................((((((((.......)))))))). (-24.49 = -24.72 + 0.23)

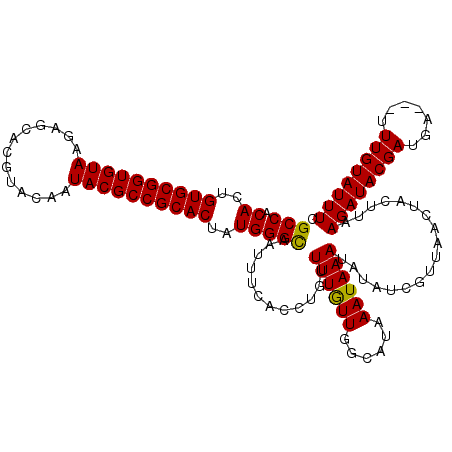
| Location | 19,019,749 – 19,019,869 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -34.23 |
| Energy contribution | -33.79 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.80 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
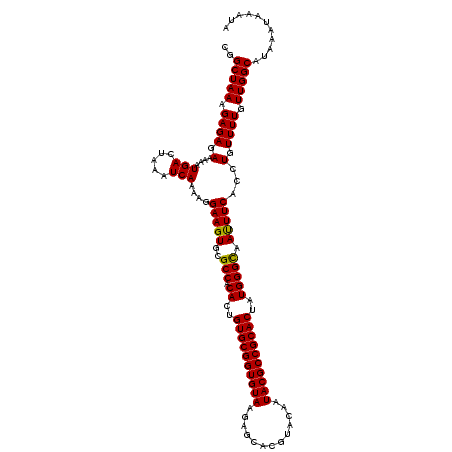
>2L_DroMel_CAF1 19019749 120 - 22407834 CGGCUAAAGAGAGAAAAAUGACUAAAUCAAAAGGAAGUGCGCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGUAAUUUCACCUGUUUUAUUGGCAUAAAUAAAUA ..(((((.((((.(....(((.....)))....(((((..(((.((..((((((((((.............))))))))))..))))).)))))...).)))).)))))........... ( -32.12) >DroSec_CAF1 23798 120 - 1 CGGCUAAAGAGAGAAAAAUGACUAAAUCAAAAGGAAGUGCGCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGCAACUUCACCUGUUUUGUUGGCAUAAAUAAAUA ..(((((.((((.(....(((.....)))....(((((..(((.((..((((((((((.............))))))))))..))))).)))))...).)))).)))))........... ( -36.02) >DroSim_CAF1 21408 120 - 1 CGGCUAAAGAGAGAAAAAUGACUAAAUCAAAAGGAAGUGCGCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGCAAUUUCACCUGUUUUGUUGGCUAAAAUAAACA .((((((.((((.(....(((.....)))....(((((..(((.((..((((((((((.............))))))))))..))))).)))))...).)))).)))))).......... ( -34.62) >consensus CGGCUAAAGAGAGAAAAAUGACUAAAUCAAAAGGAAGUGCGCCACACUGUGCGGUGUAAGAGCACGUACAAUACGCCGCACUAUGGGCAAUUUCACCUGUUUUGUUGGCAUAAAUAAAUA ..(((((.((((.(....(((.....)))....(((((..(((.((..((((((((((.............))))))))))..))))).)))))...).)))).)))))........... (-34.23 = -33.79 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:19 2006