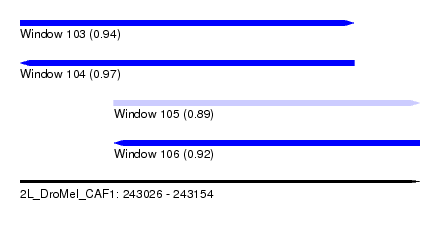
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 243,026 – 243,154 |
| Length | 128 |
| Max. P | 0.966382 |

| Location | 243,026 – 243,133 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.93 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -19.03 |
| Energy contribution | -18.79 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 243026 107 + 22407834 ACUU----------UUCGUUGCAAUAGCUCUAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUAUCA-CAGAAGGGCACAUAUAACGUACAGUUUUACCCCCCAAAAU-GUUGAGCA ....----------............((((.(((((((..((((.((((((.((((..((((....((.-.....))...))))...)))).)))))).))))..)))).-))))))). ( -27.80) >DroPse_CAF1 43144 116 + 1 AAUUGUUGUUGCGA-UUGUUGCUGUUGCAUCAAGUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUCG-AAUAUUGUCACAUAGAACGUACAGUUUGACCCCUCAAAUU-UUUGAGCA ((((((....))))-))........(((.(((((.(((.(((((..(((((.((((.(((((.......-..........)))))..)))).)))))..))))).)))..-)))))))) ( -26.73) >DroGri_CAF1 42835 109 + 1 -----GACUU-UU-UAGGUUGCUAUCACUUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUCU-U-CAUUUACACAUAUAACGUACAGUUUGACCCCCUAAACA-UUUGAGUG -----((((.-..-..)))).....(((((.((..((((.((((..(((((.((((..((((.......-.-........))))...)))).)))))..)))).))))..-)).))))) ( -22.99) >DroWil_CAF1 44371 112 + 1 A----UUGUUUACA-UUGUUGAUUUAACAUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGGAUUUCUUAAGAUUUGCACAUACAACGUACAGUUUAACCCCAUAA-AU-GUUGUGUA .----.........-...........(((.((((.((((.((((..(((((.((((..(((((...((....))....).))))...)))).)))))..)))).)))-).-))))))). ( -25.30) >DroAna_CAF1 41181 117 + 1 ACUU-UAGUUGAGAUUUGUUGACUUUGCUUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUCU-AAGAUUGCCACAUAGAACGUACAGUUUGACCCCCCAAAAGGGUUGAGCU ....-.(((..(......)..)))..(((.((((((((..((((..(((((.((((.(((((((((...-..))))....)))))..)))).)))))....))))..))))))))))). ( -28.20) >DroPer_CAF1 43601 116 + 1 AAUUGUUGUUGCGA-UUGUUGCUGUUGCAUCAAGUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUCG-AAUAUUGUCACAUAGAACGUACAGUUUGACCCCUCAAAUU-UUUGAGCA ((((((....))))-))........(((.(((((.(((.(((((..(((((.((((.(((((.......-..........)))))..)))).)))))..))))).)))..-)))))))) ( -26.73) >consensus A_UU_UUGUUGAGA_UUGUUGCUAUUGCAUCAACUUUUAAGGGGAAAAGCUAUACGCUUAUGAAUUUCU_AAGAUUGGCACAUAGAACGUACAGUUUGACCCCCCAAAAU_GUUGAGCA ..........................((.(((((.(((..((((..(((((.((((..((((..................))))...)))).)))))..))))..)))...))))))). (-19.03 = -18.79 + -0.25)

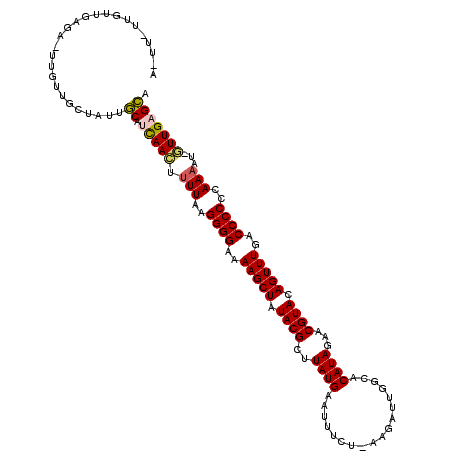
| Location | 243,026 – 243,133 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.93 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.62 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 243026 107 - 22407834 UGCUCAAC-AUUUUGGGGGGUAAAACUGUACGUUAUAUGUGCCCUUCUG-UGAUAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUAGAGCUAUUGCAACGAA----------AAGU .(((((((-.((((((((((.(((((((((((((.((((..........-.......))))))))))))).))))))))))))))))).))))............----------.... ( -34.23) >DroPse_CAF1 43144 116 - 1 UGCUCAAA-AAUUUGAGGGGUCAAACUGUACGUUCUAUGUGACAAUAUU-CGAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAACUUGAUGCAACAGCAACAA-UCGCAACAACAAUU (((.....-..(((((((((..((.(((((((((...(((((.......-......)))))))))))))).))..)))))))))..(((.(((....))).)))-..)))......... ( -31.22) >DroGri_CAF1 42835 109 - 1 CACUCAAA-UGUUUAGGGGGUCAAACUGUACGUUAUAUGUGUAAAUG-A-AGAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGUGAUAGCAACCUA-AA-AAGUC----- (((((((.-..(((((((((..((.(((((((((.((....)).(((-(-(....))))).))))))))).))..)))))))))..))).))))...........-..-.....----- ( -32.80) >DroWil_CAF1 44371 112 - 1 UACACAAC-AU-UUAUGGGGUUAAACUGUACGUUGUAUGUGCAAAUCUUAAGAAAUCCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAUGUUAAAUCAACAA-UGUAAACAA----U ((((....-.(-(((.((((..((.(((((((((.((((.(....((....))...)))))))))))))).))..)))).)))).((((((.....))))))..-)))).....----. ( -29.70) >DroAna_CAF1 41181 117 - 1 AGCUCAACCCUUUUGGGGGGUCAAACUGUACGUUCUAUGUGGCAAUCUU-AGAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGCAAAGUCAACAAAUCUCAACUA-AAGU .(((((((..((((((((((..((.(((((((((.((((..........-.......))))))))))))).))..)))))))))))))).))).....................-.... ( -32.33) >DroPer_CAF1 43601 116 - 1 UGCUCAAA-AAUUUGAGGGGUCAAACUGUACGUUCUAUGUGACAAUAUU-CGAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAACUUGAUGCAACAGCAACAA-UCGCAACAACAAUU (((.....-..(((((((((..((.(((((((((...(((((.......-......)))))))))))))).))..)))))))))..(((.(((....))).)))-..)))......... ( -31.22) >consensus UGCUCAAA_AUUUUGGGGGGUCAAACUGUACGUUCUAUGUGACAAUCUU_AGAAAUUCAUAAGCGUAUAGCUUUUCCCCUUAAAAGUUGAAGCAAAAGCAACAA_UCGCAACAA_AA_U .((((((....(((((((((..((.(((((((((.((((..................))))))))))))).))..)))))))))..)))).)).......................... (-25.90 = -25.62 + -0.28)

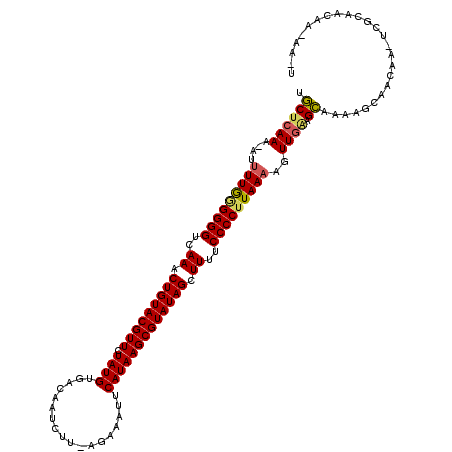
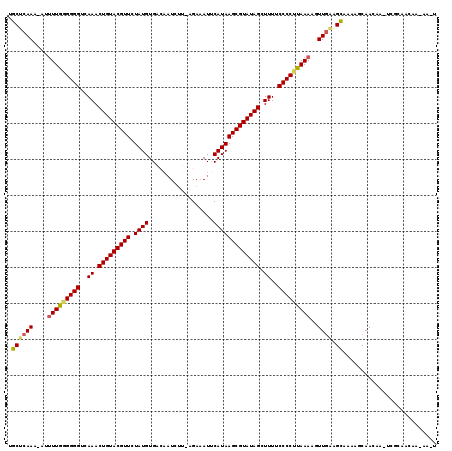
| Location | 243,056 – 243,154 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.12 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.79 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 243056 98 + 22407834 GGGGAAAAGCUAUACGCUUAUGAAUAUCA-CAGAAGGGCACAUAUAACGUACAGUUUUACCCCCCAAAAU-GUUGAGCAAUAUUGUCCAGAUU-GUACAAU--- ((((.((((((.((((..((((....((.-.....))...))))...)))).)))))).)))).......-((((.(((((.........)))-)).))))--- ( -22.10) >DroSim_CAF1 36420 98 + 1 GGGGAAAAGCUAUACGCUUAUGAAUUUCA-CAGAAGGGCACAUAUAACGUACAGUUUCACCCCCCAAAAU-GUUGAGCAAUUUUGUCCAGAUU-GUACAAG--- ((((..(((((.((((..((((..((((.-..))))....))))...)))).)))))..)))).......-.(((.(((((((.....)))))-)).))).--- ( -22.50) >DroEre_CAF1 36397 97 + 1 GGGGAAAAGCUAUACGCUUAUGAAUUUUA-CAGAAGGACACAUAUAACGUACAGUUUCACCCCCUAAAAU-GUUGAGCAAUUUUGUC-AGGUU-CUACAAG--- ((((..(((((.((((..((((..((((.-....))))..))))...)))).)))))..))))......(-((.((((.........-..)))-).)))..--- ( -21.00) >DroWil_CAF1 44406 96 + 1 GGGGAAAAGCUAUACGCUUAUGGAUUUCUUAAGAUUUGCACAUACAACGUACAGUUUAACCCCAUAA-AU-GUUGUGUAGAAUUG-----AUU-CUUUAAUUUA ((((..(((((.((((..(((((...((....))....).))))...)))).)))))..))))....-..-........((((((-----(..-..))))))). ( -20.20) >DroMoj_CAF1 40173 91 + 1 GGGGAAAAGCUAUACGCUUAUGGAUUUCA-U-UGUUUACACAUACAACGUACAGUUUGACCCCCUAAACA-UUUGAGUGUUUUGUU-CACAUUUG--------- ((((..(((((.((((..((((.......-.-........))))...)))).)))))..))))..(((((-(....))))))....-........--------- ( -19.09) >DroAna_CAF1 41220 88 + 1 GGGGAAAAGCUAUACGCUUAUGAAUUUCU-AAGAUUGCCACAUAGAACGUACAGUUUGACCCCCCAAAAGGGUUGAGCU------UCCAGAA------AAC--- ..((((((((.....))))......((((-(.(.......).)))))......(((..((((.......))))..))))------)))....------...--- ( -18.70) >consensus GGGGAAAAGCUAUACGCUUAUGAAUUUCA_CAGAAGGGCACAUAUAACGUACAGUUUCACCCCCCAAAAU_GUUGAGCAAUUUUGUCCAGAUU_GUACAAG___ ((((..(((((.((((..((((..................))))...)))).)))))..))))......................................... (-15.79 = -15.79 + -0.00)

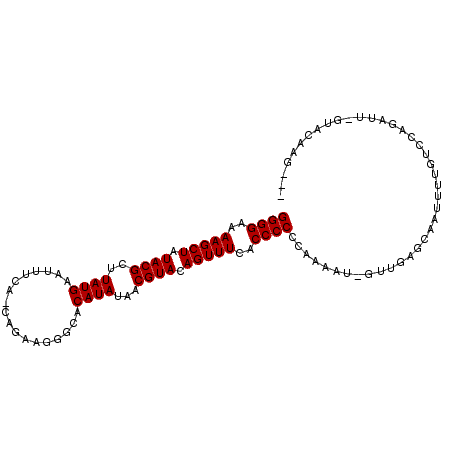
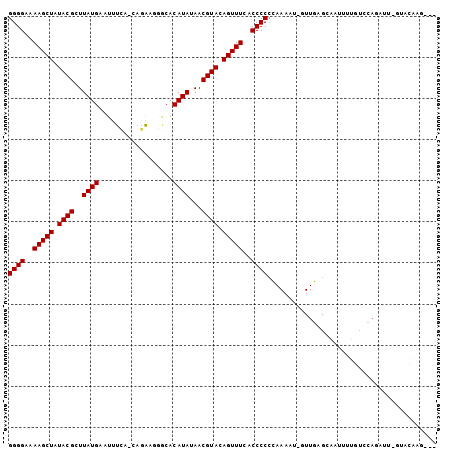
| Location | 243,056 – 243,154 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.12 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 243056 98 - 22407834 ---AUUGUAC-AAUCUGGACAAUAUUGCUCAAC-AUUUUGGGGGGUAAAACUGUACGUUAUAUGUGCCCUUCUG-UGAUAUUCAUAAGCGUAUAGCUUUUCCCC ---(((((.(-.....).)))))....(((((.-...)))))(((.(((((((((((((.((((..........-.......))))))))))))).))))))). ( -20.93) >DroSim_CAF1 36420 98 - 1 ---CUUGUAC-AAUCUGGACAAAAUUGCUCAAC-AUUUUGGGGGGUGAAACUGUACGUUAUAUGUGCCCUUCUG-UGAAAUUCAUAAGCGUAUAGCUUUUCCCC ---.......-........((((((........-)))))).((((.(((.(((((((((.((((..........-.......))))))))))))).))).)))) ( -21.63) >DroEre_CAF1 36397 97 - 1 ---CUUGUAG-AACCU-GACAAAAUUGCUCAAC-AUUUUAGGGGGUGAAACUGUACGUUAUAUGUGUCCUUCUG-UAAAAUUCAUAAGCGUAUAGCUUUUCCCC ---.((((..-.....-.))))...........-.......((((.(((.(((((((((.((((..........-.......))))))))))))).))).)))) ( -20.03) >DroWil_CAF1 44406 96 - 1 UAAAUUAAAG-AAU-----CAAUUCUACACAAC-AU-UUAUGGGGUUAAACUGUACGUUGUAUGUGCAAAUCUUAAGAAAUCCAUAAGCGUAUAGCUUUUCCCC ........((-((.-----...)))).......-..-....((((..((.(((((((((.((((.(....((....))...)))))))))))))).))..)))) ( -19.70) >DroMoj_CAF1 40173 91 - 1 ---------CAAAUGUG-AACAAAACACUCAAA-UGUUUAGGGGGUCAAACUGUACGUUGUAUGUGUAAACA-A-UGAAAUCCAUAAGCGUAUAGCUUUUCCCC ---------....((..-..))(((((......-))))).(((((.....(((((((((...(((....)))-(-((.....))).)))))))))....))))) ( -20.20) >DroAna_CAF1 41220 88 - 1 ---GUU------UUCUGGA------AGCUCAACCCUUUUGGGGGGUCAAACUGUACGUUCUAUGUGGCAAUCUU-AGAAAUUCAUAAGCGUAUAGCUUUUCCCC ---...------..(..((------((.......))))..)((((..((.(((((((((.((((..........-.......))))))))))))).))..)))) ( -22.13) >consensus ___AUUGUAC_AAUCUGGACAAAAUUGCUCAAC_AUUUUAGGGGGUCAAACUGUACGUUAUAUGUGCCAAUCUG_UGAAAUUCAUAAGCGUAUAGCUUUUCCCC .........................................((((..((.(((((((((.((((..................))))))))))))).))..)))) (-18.22 = -18.22 + 0.00)

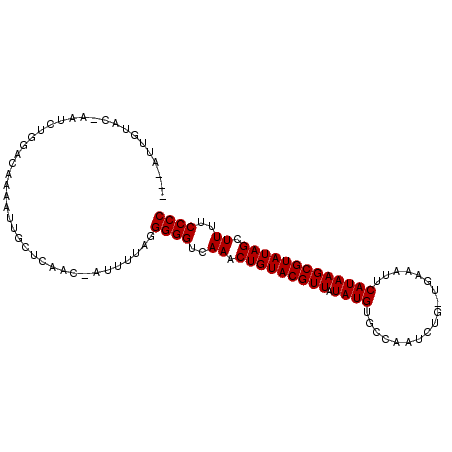
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:57 2006