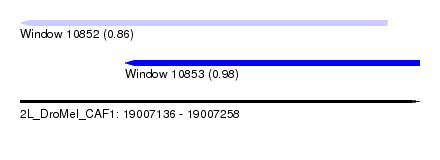
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,007,136 – 19,007,258 |
| Length | 122 |
| Max. P | 0.979525 |

| Location | 19,007,136 – 19,007,248 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -48.41 |
| Consensus MFE | -26.62 |
| Energy contribution | -27.63 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
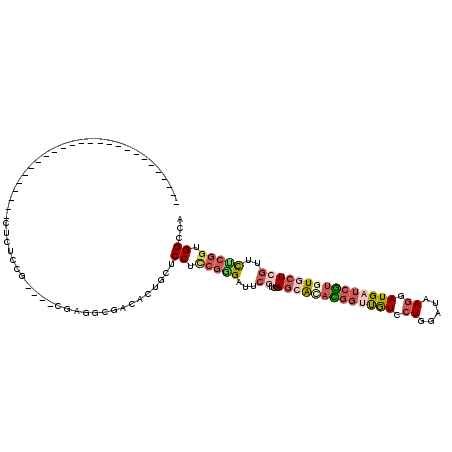
>2L_DroMel_CAF1 19007136 112 - 22407834 GGCGACACUGCUCCUCCAGGAUUCGUCGAGCACACGGUUGUCCUGGAUCAGAAUGAUCGUGUGCUCGUUCUCGGUGGCCACCAUCGUCUCCUGGCGGCGGGUGCGCAGCCGC .(((.((((.(.((.((((((.....((((((((((((..(.(((...))).)..))))))))))))....((((((...))))))..)))))).)).))))))))...... ( -53.80) >DroPse_CAF1 9200 112 - 1 CGAUGAGCUAUUCCUACGCGCCUCAUCUAAAAUAUUCGUAUCCUGUACGAGUAUGAUCAUGUGCUCGUUCUCAGCGGCCACCAGAAUCUCACUGUUGAGGGUUCGCAUCCGC .(((((((...........).))))))....((((((((((...))))))))))......((((..(..((((((((..............))))))))..)..)))).... ( -29.54) >DroSec_CAF1 10055 112 - 1 GGCGACACUGCUCCUCCGGGAUUCGUCGAGCAUACGGUUGUCCUGGAUCAGAAUGAUCGUGUGCUCGUUCUCGGUGGCCACCAUCGUCUCCUGGCGGCGGGUGCGCAGCCGC .(((...((((.((.((((((.....((((((((((((..(.(((...))).)..)))))))))))).)))))).)).((((.(((((....)))))..)))).)))).))) ( -52.50) >DroEre_CAF1 10207 112 - 1 GGCCGCACUGUUCCUCCGGGAUUCGUCCAGCACAUGGUUGUCCUGGAUAAGGAUGAUCGUGUGCUCGUUCCCGGUGGCCACCAGCGUCUCCUGGCGGCGGGUGCGCAGCCGC (((((((((....(.((((((..((...((((((((((..((((.....))))..)))))))))))).)))))).)(((.((((......)))).))).))))))..))).. ( -62.40) >DroYak_CAF1 7280 112 - 1 AGCGACACUGUUCCUCCGGGAUUCGUCUAGCACACGGUUGUCCUGGAUAAGGAUGAUCGUAUGCUCGUGCCCGGUGGCCACCAUCGUCUCCUGGCGGCGGGUGCGCAGCCGC .(((.(((....((.(((((...((...((((.(((((..((((.....))))..))))).))))))..))))).))...((..((((....))))..))))))))...... ( -50.80) >DroAna_CAF1 1962 112 - 1 AGCGGAAGUGCUCCGUCGAGUCUGGUCCAGGGACUGGACAUCCUGGAUGACGAUGAUCGUGUGCUGGCUUUCGGUGGCCACCAUCAGCUCCUGUUUGCGUGUCCGCAAACGA .(((((.....))))).(((.(((((((((((........)))))))).(((.....)))(((.(((((......))))).))))))))).((((((((....)))))))). ( -41.40) >consensus GGCGACACUGCUCCUCCGGGAUUCGUCCAGCACACGGUUGUCCUGGAUAAGGAUGAUCGUGUGCUCGUUCUCGGUGGCCACCAUCGUCUCCUGGCGGCGGGUGCGCAGCCGC .(((...((((.((.(((((...((...(((((((((((((.((.....)).)))))))))))))))..))))).)).......((((....))))))))...)))...... (-26.62 = -27.63 + 1.01)



| Location | 19,007,168 – 19,007,258 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.87 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -17.45 |
| Energy contribution | -18.93 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19007168 90 - 22407834 --------------------------CUCUCCG----CGAGGCGACACUGCUCCUCCAGGAUUCGUCGAGCACACGGUUGUCCUGGAUCAGAAUGAUCGUGUGCUCGUUCUCGGUGGCCA --------------------------.((.(((----.(((((((..(((......)))...)))))(((((((((((..(.(((...))).)..)))))))))))..)).))).))... ( -37.90) >DroPse_CAF1 9232 120 - 1 GUCACAUUGCAUACAGAAUUCAUUCACUCUUCUUGGACGACGAUGAGCUAUUCCUACGCGCCUCAUCUAAAAUAUUCGUAUCCUGUACGAGUAUGAUCAUGUGCUCGUUCUCAGCGGCCA (((.((((((.....(((....)))..(((....))).).))))).(((......(((.((..(((.....((((((((((...))))))))))....))).)).)))....)))))).. ( -24.30) >DroSec_CAF1 10087 90 - 1 --------------------------CUCUCCG----CGAGGCGACACUGCUCCUCCGGGAUUCGUCGAGCAUACGGUUGUCCUGGAUCAGAAUGAUCGUGUGCUCGUUCUCGGUGGCCA --------------------------.((.((.----...)).)).......((.((((((.....((((((((((((..(.(((...))).)..)))))))))))).)))))).))... ( -39.60) >DroEre_CAF1 10239 90 - 1 --------------------------AGCGCCG----CGAGGCCGCACUGUUCCUCCGGGAUUCGUCCAGCACAUGGUUGUCCUGGAUAAGGAUGAUCGUGUGCUCGUUCCCGGUGGCCA --------------------------.(((((.----...)).)))......((.((((((..((...((((((((((..((((.....))))..)))))))))))).)))))).))... ( -48.90) >DroYak_CAF1 7312 90 - 1 --------------------------ACCGCCG----GGAAGCGACACUGUUCCUCCGGGAUUCGUCUAGCACACGGUUGUCCUGGAUAAGGAUGAUCGUAUGCUCGUGCCCGGUGGCCA --------------------------.((((((----((..(((((...(((((...)))))..))).((((.(((((..((((.....))))..))))).)))).)).))))))))... ( -42.70) >DroAna_CAF1 1994 83 - 1 -------------------------------------CGAAGCGGAAGUGCUCCGUCGAGUCUGGUCCAGGGACUGGACAUCCUGGAUGACGAUGAUCGUGUGCUGGCUUUCGGUGGCCA -------------------------------------((((((...((..(((((..(((((..(((....)))..))).)).))))..(((.....))))..)).)).))))....... ( -27.70) >consensus __________________________CUCUCCG____CGAGGCGACACUGCUCCUCCGGGAUUCGUCCAGCACACGGUUGUCCUGGAUAAGGAUGAUCGUGUGCUCGUUCUCGGUGGCCA ....................................................((.(((((...((...(((((((((((((.((.....)).)))))))))))))))..))))).))... (-17.45 = -18.93 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:13 2006