
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,004,871 – 19,005,070 |
| Length | 199 |
| Max. P | 0.999583 |

| Location | 19,004,871 – 19,004,991 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -51.86 |
| Consensus MFE | -48.74 |
| Energy contribution | -49.34 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19004871 120 - 22407834 GGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAACUGGUGCUCAUCGCCAGCAACACGCCCGCCCUGAGGUGAGA (((((((((((((((((((((((.....)))).))))).((((.....))))........((((((((((...)))...)))))))))))))))))).....)))((((....))))... ( -53.50) >DroSec_CAF1 7865 120 - 1 GGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAACUGGUGCUCAUCGCCAGCAACACGCCCGCCCUGAGGUGAGA (((((((((((((((((((((((.....)))).))))).((((.....))))........((((((((((...)))...)))))))))))))))))).....)))((((....))))... ( -53.50) >DroSim_CAF1 6223 120 - 1 GGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAACUGGUGCUCAUCGCCAGCAACACGCCCGCCCUGAGGUGAGA (((((((((((((((((((((((.....)))).))))).((((.....))))........((((((((((...)))...)))))))))))))))))).....)))((((....))))... ( -53.50) >DroEre_CAF1 7983 120 - 1 GGCUCUGGUGAUGAAGUCCGGCAAGUACUGCCUGGGCUACAAACAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAACUGGUGCUCAUCGCCAGCAACACCCCCGCCCUGAGGUGAGA (...(((((((((((((((((((.....)))).)))))......................((((((((((...)))...)))))))))))))))))...).....((((....))))... ( -46.00) >DroYak_CAF1 5030 120 - 1 GGCGCUGGUGAUGAAGUCCGGCAAGUACUGCCUGGGAUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAACUGGUGCUCAUCGCCAGCAACACCCCCGCCCUGAGGUGAGA ...(((((((((((.(((...((((..((((.((.....)).))))..))))..)))...((((((((((...)))...))))))))))))))))))..((((.(......).))))... ( -52.80) >consensus GGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAACUGGUGCUCAUCGCCAGCAACACGCCCGCCCUGAGGUGAGA ...((((((((((((((((((((.....)))).))))).((((.....))))........((((((((((...)))...))))))))))))))))))........((((....))))... (-48.74 = -49.34 + 0.60)


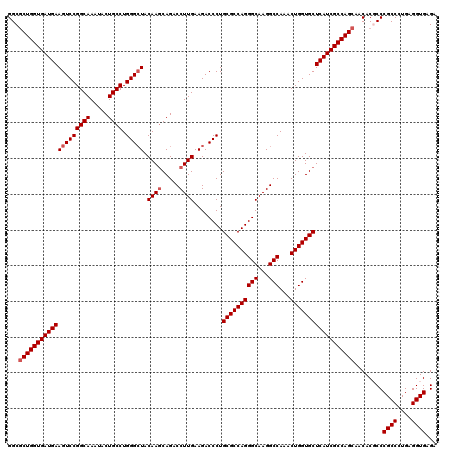
| Location | 19,004,911 – 19,005,030 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -45.56 |
| Consensus MFE | -39.60 |
| Energy contribution | -39.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19004911 119 + 22407834 GUUUGGCCUUGCCCUGGCGCAGGGUCUUCAAGGUCUGCUUGUAGCCCAGGCAGUAUUUGCCGGACUUCAUCACCAGCGCCAGACGG-GCGUUGGUGCUCUCCAGAGCCUUCUUUUGUUUC (((((((...((((((...)))))).....(((((((((((.....))))))).)))))))))))......(((((((((.....)-))))))))((((....))))............. ( -47.20) >DroSec_CAF1 7905 120 + 1 GUUUGGCCUUGCCCUGGCGCAGGGUCUUCAAGGUCUGCUUGUAGCCCAGGCAGUAUUUGCCGGACUUCAUCACCAGCGCCAGACGGGGCGUUGGUGCUCUCCAGAGCCUUCUUUUGUUUC (((((((...((((((...)))))).....(((((((((((.....))))))).)))))))))))......(((((((((......)))))))))((((....))))............. ( -47.40) >DroSim_CAF1 6263 119 + 1 GUUUGGCCUUGCCCUGGCGCAGGGUCUUCAAGGUCUGCUUGUAGCCCAGGCAGUAUUUGCCGGACUUCAUCACCAGCGCCAGACGG-GCGUUGGUGCUCUCCAGAGCCUUCUUUUGUUUC (((((((...((((((...)))))).....(((((((((((.....))))))).)))))))))))......(((((((((.....)-))))))))((((....))))............. ( -47.20) >DroEre_CAF1 8023 119 + 1 GUUUGGCCUUGCCCUGGCGCAGGGUCUUCAAGGUCUGUUUGUAGCCCAGGCAGUACUUGCCGGACUUCAUCACCAGAGCCAGACGG-GCAUUGGUGCUCUCCAGCGCCUUCUUUUGUUUC ....(((...)))..(((((.((((((.((((..(((((((.....)))))))..))))..)))).....((((((.(((.....)-)).))))))....)).)))))............ ( -41.70) >DroYak_CAF1 5070 119 + 1 GUUUGGCCUUGCCCUGGCGCAGGGUCUUCAAGGUCUGCUUGUAUCCCAGGCAGUACUUGCCGGACUUCAUCACCAGCGCCAAACGG-GCAUUGGUGCUCUCCAACGCCUUCUUUUGUUUC ....(((..(((((((((((.((((((.((((..(((((((.....)))))))..))))..)))).......)).))))))...))-)))((((......)))).)))............ ( -44.31) >consensus GUUUGGCCUUGCCCUGGCGCAGGGUCUUCAAGGUCUGCUUGUAGCCCAGGCAGUAUUUGCCGGACUUCAUCACCAGCGCCAGACGG_GCGUUGGUGCUCUCCAGAGCCUUCUUUUGUUUC (((((((...((((((...)))))).....(((((((((((.....))))))).))))))))))).....(((((((((........)))))))))........................ (-39.60 = -39.60 + 0.00)

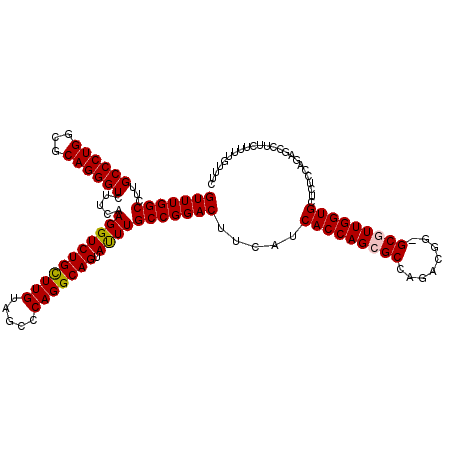
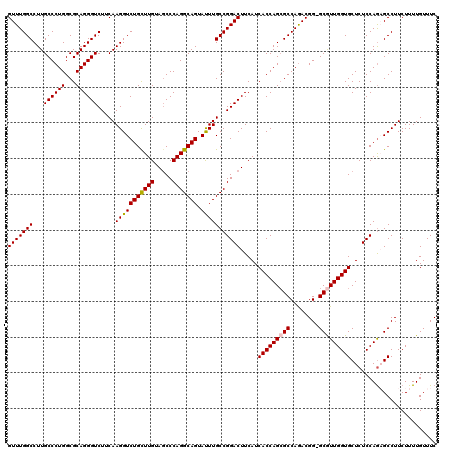
| Location | 19,004,911 – 19,005,030 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -44.36 |
| Consensus MFE | -37.56 |
| Energy contribution | -37.68 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19004911 119 - 22407834 GAAACAAAAGAAGGCUCUGGAGAGCACCAACGC-CCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAAC ............((((.(((......)))..((-((...((((((.(((.....(((((((((.....)))).))))).((((.....))))...)))..))))))))))..)))).... ( -44.60) >DroSec_CAF1 7905 120 - 1 GAAACAAAAGAAGGCUCUGGAGAGCACCAACGCCCCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAAC ............(((((((.....(((((.((((......)))).))))).((.(((((((((.....)))).))))).))..)))).((((....(((((...)))))))))))).... ( -44.70) >DroSim_CAF1 6263 119 - 1 GAAACAAAAGAAGGCUCUGGAGAGCACCAACGC-CCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAAC ............((((.(((......)))..((-((...((((((.(((.....(((((((((.....)))).))))).((((.....))))...)))..))))))))))..)))).... ( -44.60) >DroEre_CAF1 8023 119 - 1 GAAACAAAAGAAGGCGCUGGAGAGCACCAAUGC-CCGUCUGGCUCUGGUGAUGAAGUCCGGCAAGUACUGCCUGGGCUACAAACAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAAC ............((((.(((......))).)))-).((((.(((((((((.((.(((((((((.....)))).))))).))..(((............)))))))))))).))))..... ( -42.90) >DroYak_CAF1 5070 119 - 1 GAAACAAAAGAAGGCGUUGGAGAGCACCAAUGC-CCGUUUGGCGCUGGUGAUGAAGUCCGGCAAGUACUGCCUGGGAUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAAC ............((((((((......)))))))-).((((((((((((.........)))))..((.((((.((.....)).))))))((((....(((((...)))))))))))))))) ( -45.00) >consensus GAAACAAAAGAAGGCUCUGGAGAGCACCAACGC_CCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUACAAGCAGACCUUGAAGACCCUGCGCCAGGGCAAGGCCAAAC ............(((.(((.....(((((.(((........))).))))).((.(((((((((.....)))).))))).))..)))..((((....(((((...)))))))))))).... (-37.56 = -37.68 + 0.12)

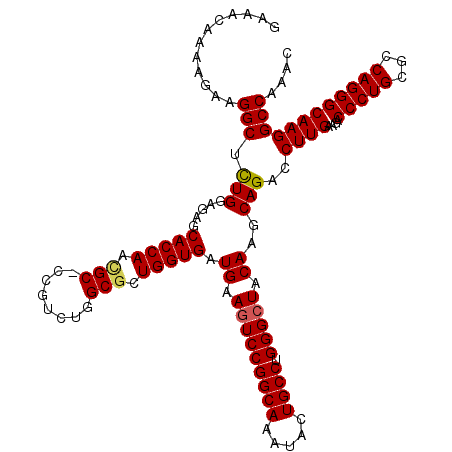
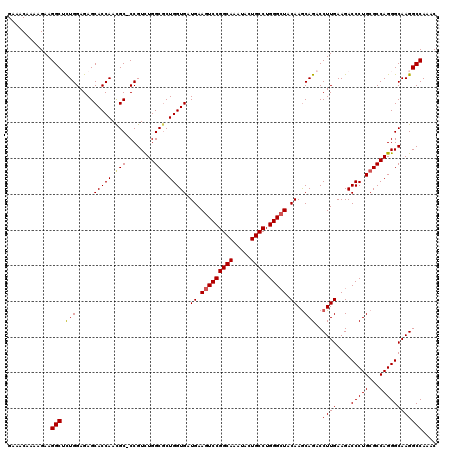
| Location | 19,004,951 – 19,005,070 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.16 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
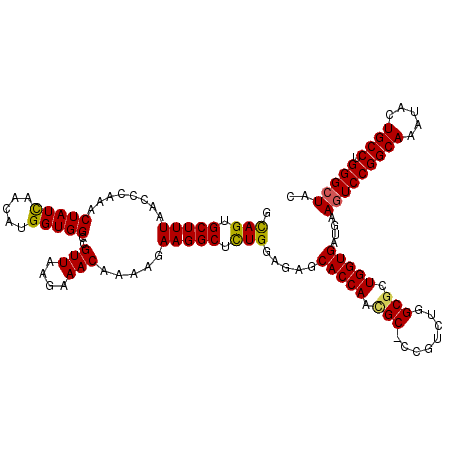
>2L_DroMel_CAF1 19004951 119 - 22407834 GCAGUGCUUUAACCCAAACUAUCAACAUGGUGGCCGUUAAGAAACAAAAGAAGGCUCUGGAGAGCACCAACGC-CCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUAC .(((.(((((........(((((.....)))))..(((....))).....))))).))).....(((((.(((-(.....)))).)))))....(((((((((.....)))).))))).. ( -38.50) >DroSec_CAF1 7945 120 - 1 AUAGUGCUUUAACCCAAACUAUUAUCAUGGUGGCCGUUAAGAAACAAAAGAAGGCUCUGGAGAGCACCAACGCCCCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUAC ...((..((((((....(((((....)))))....))))))..))........((((....))))((((.((((......)))).)))).....(((((((((.....)))).))))).. ( -36.80) >DroSim_CAF1 6303 119 - 1 ACAGUGCUUUAACCCAAACUAUUAUCAUGGUGGCCGUUAAGAAACAAAAGAAGGCUCUGGAGAGCACCAACGC-CCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUAC .(((.(((((...((..(((((....)))))))..(((....))).....))))).))).....(((((.(((-(.....)))).)))))....(((((((((.....)))).))))).. ( -37.30) >DroEre_CAF1 8063 119 - 1 GCAGUGCUUUAACCCAAACUAUCAACAUGGUGGCCGUUAAGAAACAAAAGAAGGCGCUGGAGAGCACCAAUGC-CCGUCUGGCUCUGGUGAUGAAGUCCGGCAAGUACUGCCUGGGCUAC .(((((((((........(((((.....)))))..(((....))).....))))))))).....(((((..((-(.....)))..)))))....(((((((((.....)))).))))).. ( -39.80) >DroYak_CAF1 5110 119 - 1 GCAGUGCUUUAACCCAAACUAUCAACAUGGUGGCCGUUAAGAAACAAAAGAAGGCGUUGGAGAGCACCAAUGC-CCGUUUGGCGCUGGUGAUGAAGUCCGGCAAGUACUGCCUGGGAUAC (((((((((....((((((..((...((((...))))...))..........((((((((......)))))))-).)))))).(((((.........))))))))))))))......... ( -39.90) >consensus GCAGUGCUUUAACCCAAACUAUCAACAUGGUGGCCGUUAAGAAACAAAAGAAGGCUCUGGAGAGCACCAACGC_CCGUCUGGCGCUGGUGAUGAAGUCCGGCAAAUACUGCCUGGGCUAC .(((.(((((........(((((.....)))))..(((....))).....))))).))).....(((((.(((........))).)))))....(((((((((.....)))).))))).. (-31.44 = -31.16 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:08 2006