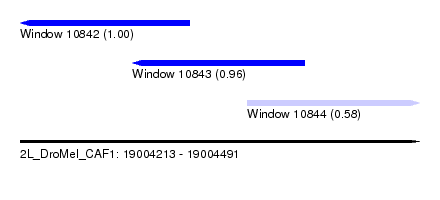
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,004,213 – 19,004,491 |
| Length | 278 |
| Max. P | 0.998406 |

| Location | 19,004,213 – 19,004,331 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.79 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19004213 118 - 22407834 CAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUUAAUCGAACAAGACGACACAAUAAAGUUUUGUAUGAGCAGAGAUAAAGUUUAAAAUAAAAACACUAUUGUU-UUGGUAAA ......((....((((....)))).(((((.(((((((((..(((.(((((((...........))))))).)))..)))))))))))))).....((((((....))))-)))).... ( -28.10) >DroSec_CAF1 7209 118 - 1 CAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUUAAUCGAACAAGACGACACAAUAAAGUUUUGUAUGAGCAGAGAUAAAGUUUAAAAUAAAAACACUAUUAUU-UUGGUAAA ............((((....)))).(((((.(((((((((..(((.(((((((...........))))))).)))..))))))))))))))..........((((.....-.))))... ( -26.80) >DroSim_CAF1 5567 119 - 1 CAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUUAAUCGAACAAGACGACACAAUAAAGUUUUGUAUGAGCAGAGAUAAAGUUUAAAAUAAAAACACUAUUAUUUUUGGUAAA .((((.......((((....)))).(((((.(((((((((..(((.(((((((...........))))))).)))..)))))))))))))).....................))))... ( -26.50) >DroEre_CAF1 7315 116 - 1 CAUCAUCCGUUCGCUGGAGUCGGCCUAAAC--UUAUUUUUAAUCGAACAAGACGACACAAUAAAGUUUUGUAUGAGCAGAGAUAAAGUUUAAAAUAAAAACCCUAUUAUU-UUGGUAAA .....((((.....))))....(((.((((--((((((((..(((.(((((((...........))))))).)))..)))))).))))))(((((((........)))))-)))))... ( -21.80) >DroYak_CAF1 4355 116 - 1 CAUCAUCCGCUCGCUGGAGUCGGCCUAAAC--UUAUUUUUAAUCGAACAAGACGACCCAAUAAAGUUUUGUAUGAGCAGAGAUAAAGUUUAAAAUAAAAACACUAUUAUU-UUGGUAAA ......((((((....))).)))..(((((--((((((((..(((.(((((((...........))))))).)))..)))))).)))))))..........((((.....-.))))... ( -24.40) >consensus CAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUUAAUCGAACAAGACGACACAAUAAAGUUUUGUAUGAGCAGAGAUAAAGUUUAAAAUAAAAACACUAUUAUU_UUGGUAAA .((((.......((((....)))).(((((.(((((((((..(((.(((((((...........))))))).)))..)))))))))))))).....................))))... (-23.22 = -23.62 + 0.40)

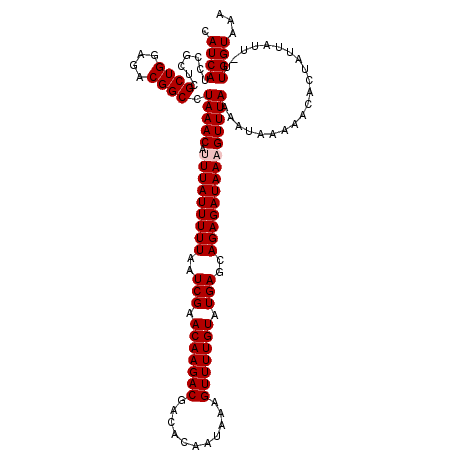
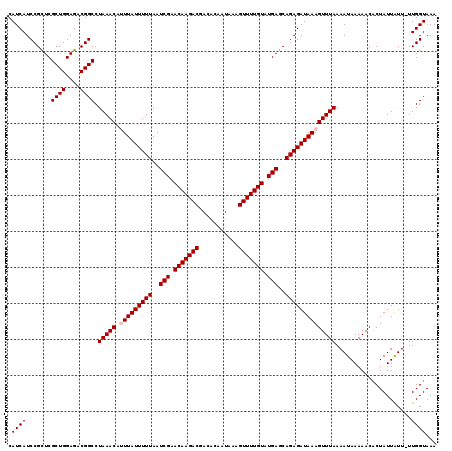
| Location | 19,004,291 – 19,004,411 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.16 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -35.82 |
| Energy contribution | -35.50 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19004291 120 - 22407834 ACCAACAUCGAGCUGGGCACCGCCUGUGGUAAAUACUUCCGCGUGUGCACCCUGUCCAUCACCGAUCCUGGAGAUUCGGACAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUU .........((((.(((((((((....(((....)))...))).)))).)).((((((((.(((....))).)))..)))))......))))((((....))))................ ( -37.30) >DroSec_CAF1 7287 120 - 1 ACCAACAUCGAGCUGGGCACCGCCUGCGGUAAAUACUUCCGCGUGUGCACCCUGUCCAUCACCGAUCCUGGAGAUUCGGACAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUU .........((((.(((...(((.(((((.........))))).)))..)))((((((((.(((....))).)))..)))))......))))((((....))))................ ( -38.10) >DroSim_CAF1 5646 120 - 1 ACCAACAUCGAGCUGGGCACCGCCUGCGGUAAAUACUUCCGCGUGUGCACCCUGUCCAUCACCGAUCCUGGAGAUUCGGACAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUU .........((((.(((...(((.(((((.........))))).)))..)))((((((((.(((....))).)))..)))))......))))((((....))))................ ( -38.10) >DroEre_CAF1 7393 118 - 1 ACCAACAUCGAGCUGGGCACCGCCUGCGGUAAAUAUUUCCGCGUGUGCACCCUGUCCAUCACCGAUCCUGGAGAUUCGGACAUCAUCCGUUCGCUGGAGUCGGCCUAAAC--UUAUUUUU .........(..(.(((...(((.(((((.........))))).)))..))).)..)....((((..(..(.((..((((.....)))).)).)..)..)))).......--........ ( -33.20) >DroYak_CAF1 4433 118 - 1 ACCAACAUUGAGCUGGGCACCGCCUGCGGUAAAUACUUCCGCGUGUGCACCCUGUCCAUCACCGAUCCUGGAGAUUCGGACAUCAUCCGCUCGCUGGAGUCGGCCUAAAC--UUAUUUUU ........((((.(((((((((....))))....(((((.(((.(((.....((((((((.(((....))).)))..))))).....))).))).)))))..)))))..)--)))..... ( -36.40) >consensus ACCAACAUCGAGCUGGGCACCGCCUGCGGUAAAUACUUCCGCGUGUGCACCCUGUCCAUCACCGAUCCUGGAGAUUCGGACAUCAUCCGCUCGCUGGAGACGGCCUAAACAUUUAUUUUU .........((((.(((...(((.(((((.........))))).)))..)))((((((((.(((....))).)))..)))))......))))((((....))))................ (-35.82 = -35.50 + -0.32)

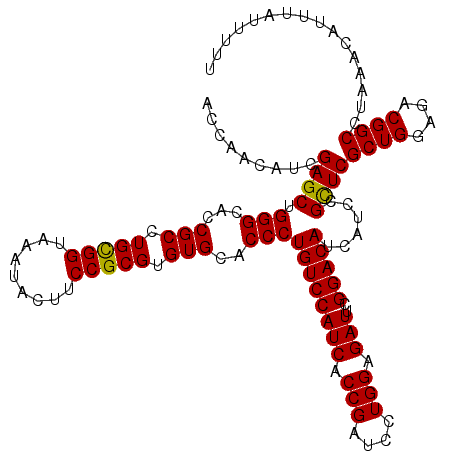

| Location | 19,004,371 – 19,004,491 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -42.04 |
| Energy contribution | -41.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19004371 120 + 22407834 GGAAGUAUUUACCACAGGCGGUGCCCAGCUCGAUGUUGGUGCCGCUGUAGUGCUGGACUUCAGUCUUGGCCAGCAUAGCGUAGUACUCGAUCUCGGACUUCCUGCAAUGUGAAGAGCACA ((((((...(((.((.(((((..((.(((.....)))))..)))))((.(((((((.((........))))))))).)))).))).(((....))))))))).....((((.....)))) ( -44.50) >DroSec_CAF1 7367 120 + 1 GGAAGUAUUUACCGCAGGCGGUGCCCAGCUCGAUGUUGGUGCCGCUGUAGUGCUGGACUUCAGUCUUGGCCAGCAUGGCGUAGUACUCGAUCUCGGACUUCCUGCAAUGGGAAGAGCACA .((.(((((...(((.(((((..((.(((.....)))))..)))))...(((((((.((........))))))))).))).))))))).........((((((.....))))))...... ( -45.10) >DroSim_CAF1 5726 120 + 1 GGAAGUAUUUACCGCAGGCGGUGCCCAGCUCGAUGUUGGUGCCGCUGUAGUGCUGGACUUCAGUCUUGGCCAGCAUGGCGUAGUACUCGAUCUCGGACUUCCUGCAAUGGGAAGAGCACA .((.(((((...(((.(((((..((.(((.....)))))..)))))...(((((((.((........))))))))).))).))))))).........((((((.....))))))...... ( -45.10) >DroEre_CAF1 7471 120 + 1 GGAAAUAUUUACCGCAGGCGGUGCCCAGCUCGAUGUUGGUGCCGCUGUAGUGCUGGACUUCAGUCUUGGCCAGCAUGGCGUAGUACUCGAUCUCGGACUUCCUGCAAUGGGAAGAGCGCA .............((((.(((..((.(((.....)))))..))))))).(((((((.((........))))))))).((((.....(((....))).((((((.....)))))).)))). ( -44.50) >DroYak_CAF1 4511 120 + 1 GGAAGUAUUUACCGCAGGCGGUGCCCAGCUCAAUGUUGGUGCCGCUGUAAUGCUGGACUUCAGUCUUGGCCAGCAUGGCGUAGUACUCGAUCUCGGACUUCCUGCAAUGGGAAAAACGGA .((.(((((...(((.(((((..((.(((.....)))))..)))))...(((((((.((........))))))))).))).)))))))....(((...(((((.....)))))...))). ( -43.80) >consensus GGAAGUAUUUACCGCAGGCGGUGCCCAGCUCGAUGUUGGUGCCGCUGUAGUGCUGGACUUCAGUCUUGGCCAGCAUGGCGUAGUACUCGAUCUCGGACUUCCUGCAAUGGGAAGAGCACA ((((((...(((.((.(((((..((.(((.....)))))..)))))((.(((((((.((........))))))))).)))).))).(((....))))))))).................. (-42.04 = -41.92 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:05 2006