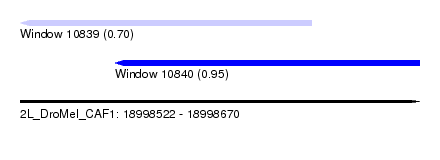
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,998,522 – 18,998,670 |
| Length | 148 |
| Max. P | 0.951810 |

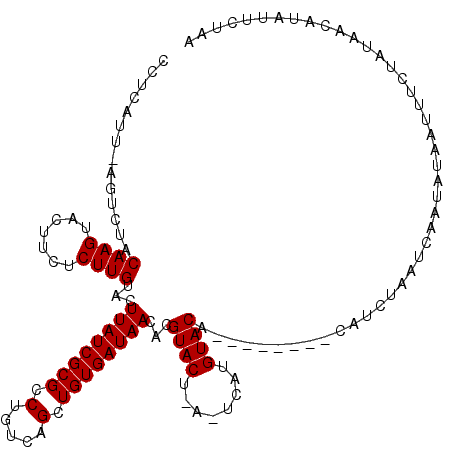
| Location | 18,998,522 – 18,998,630 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -16.77 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18998522 108 - 22407834 CCUCAUUAAGUCUACAAGUACUACUCUUGUCCUUAUCGCGCCUGUCAGCUGUGAUAACACGUAC----UCAUGUACA--------CAUCUAAUAAAUAUAAUUUCUAUAACGUAUUCCAA .........((.(((((((((.((....))..(((((((((......)).)))))))...))))----)..)))).)--------).................................. ( -14.60) >DroSim_CAF1 14595 111 - 1 CCUCAUU-AGUCUACAAGUACUUCUCUUGUACUUAUCGCGCCUGUCAGCUGUGAUAACACGUACUCACUCAUGUACA--------CAUCUAAUCAAUAUCAUUUCUAUAACAUAUUCUAA ....(((-((..((((((.......)))))).(((((((((......)).)))))))...((((........)))).--------...)))))........................... ( -16.70) >DroYak_CAF1 15068 119 - 1 CCUCAUU-AGUCUACAAGUACUUCACUUGAACUUAUCGCGCCUGUCAGAUGUGAUAACACGUACUUAUGUAUGUACAUAUACAUAUAUCUAAUCAAUACAAUUUCUCUAACAUAUUCUAA ....(((-((.....(((((((((....))).((((((((.(.....).))))))))...))))))(((((((((....))))))))))))))........................... ( -19.00) >consensus CCUCAUU_AGUCUACAAGUACUUCUCUUGUACUUAUCGCGCCUGUCAGCUGUGAUAACACGUACU_A_UCAUGUACA________CAUCUAAUCAAUAUAAUUUCUAUAACAUAUUCUAA ..............((((.......))))...((((((((.(.....).))))))))...((((........))))............................................ (-11.63 = -11.63 + 0.00)


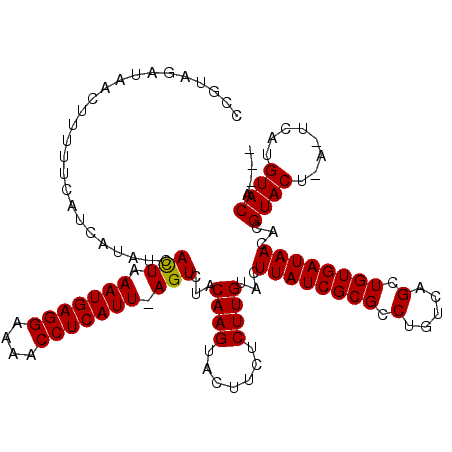
| Location | 18,998,557 – 18,998,670 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -21.02 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18998557 113 - 22407834 CCAUAGAUAUCUUAUUCAUCAUAUCACUAAAUGAGGAAAACCUCAUUAAGUCUACAAGUACUACUCUUGUCCUUAUCGCGCCUGUCAGCUGUGAUAACACGUAC----UCAUGUACA--- .....(((((..........)))))(((.(((((((....))))))).)))..(((((.......)))))..(((((((((......)).)))))))...((((----....)))).--- ( -25.40) >DroSim_CAF1 14630 116 - 1 CCGUAGAUAACUUUUUCAUCAUAUCACUCAAUGAGGAAAACCUCAUU-AGUCUACAAGUACUUCUCUUGUACUUAUCGCGCCUGUCAGCUGUGAUAACACGUACUCACUCAUGUACA--- ..((((((.....................(((((((....)))))))-.))))))((((((.......))))))(((((((......)).))))).....((((........)))).--- ( -25.85) >DroYak_CAF1 15108 118 - 1 CGGUAGACAACUUUUUCAUCGUAUUAUUAAAUGAGGA-AGCCUCAUU-AGUCUACAAGUACUUCACUUGAACUUAUCGCGCCUGUCAGAUGUGAUAACACGUACUUAUGUAUGUACAUAU ..((((((.((.........)).......(((((((.-..)))))))-.))))))(((((((((....))).((((((((.(.....).))))))))...))))))((((....)))).. ( -26.10) >consensus CCGUAGAUAACUUUUUCAUCAUAUCACUAAAUGAGGAAAACCUCAUU_AGUCUACAAGUACUUCUCUUGUACUUAUCGCGCCUGUCAGCUGUGAUAACACGUACU_A_UCAUGUACA___ .........................(((.(((((((....))))))).)))...((((.......))))...((((((((.(.....).))))))))...((((........)))).... (-21.02 = -20.80 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:01 2006