
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,984,856 – 18,985,032 |
| Length | 176 |
| Max. P | 0.664026 |

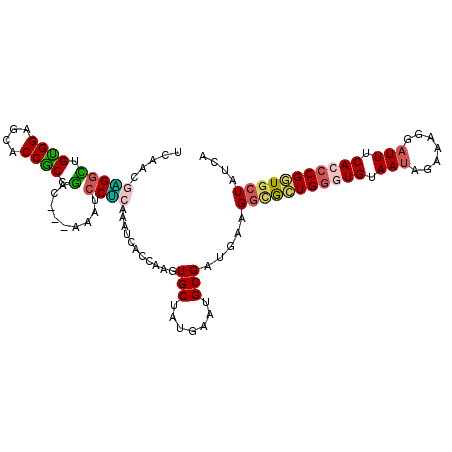
| Location | 18,984,856 – 18,984,961 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.03 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18984856 105 - 22407834 UCAGCGAGGCUGUGGAGCACCGCCGGA---AAAUCCUCAAAUCACCAAGUGCUAUGAAUGCGAUGAAGGCGCUGGGUGUAAUAGUAAGGAUUUCACCCGGUGCUACCA .((((...))))((((((((((..(..---(((((((.....((((.((((((..............)))))).))))........)))))))..).))))))).))) ( -37.66) >DroSim_CAF1 1084 105 - 1 UCAACGAGGCUGCGGAGCACCGCCGGC---AAAUCCUCAAAUCACCAAGUGCUAUGAAUGCGAUGAAGGCGCUGGGUGUAAUAGUAAGGAUUUCACCCGGUGCUACCA .............(((((((((...(.---(((((((.....((((.((((((..............)))))).))))........))))))))...))))))).)). ( -35.16) >DroEre_CAF1 1100 105 - 1 UCAACGAGGUUGUGGAGCACCGCCGAC---AAAUCCUCAAAUCACUAAGUGCUAUGAAUGCGAUGAAGGCACUGGGUGUAAUAGUAAAGAUUUCACCCGGUGCUAUCA .....((((((((.(.((...))).))---))..))))...........(((.......))).(((.(((((((((((.(((.......))).))))))))))).))) ( -33.70) >DroWil_CAF1 950 108 - 1 ACGUCGUGGUUGUGGUGCCCCACCGAGUGGCGAGGCGAACAUUCGCAAUUGCUAUGAAUGCGAUGAGGAUGGUGGAUGCAAUGAAUUGGAUUUCACACGUUGCUAUCG .((((.(.(((((((((...))))..(((((((.((((....))))..)))))))....))))).).))))...(((((((((...((.......)))))))).))). ( -33.50) >DroYak_CAF1 1077 105 - 1 UCAACGGGGCUGUGGAGCACCGCCGAC---CAAUCCUCAAAUCACCAAGUGCUAUGAAUGCGAUGAUGGCGCUGGGUGUAAUAGAAAGGAUUUCACCCGGUGCUAUCA .....(((((.(((...))).)))((.---......))......))...(((.......))).(((((((((((((((.(((.......))).))))))))))))))) ( -35.00) >DroAna_CAF1 974 102 - 1 CCAGCGGGGAUGCGGAGCACCGCCUAG---UAAUCCCGGCAUUCUCAACUGCUACGAAUGCGAC---GGGACUGGAUGUAACAGAAAAGAUUUCACCCGCUGCUAUCG .(((((((((.((((....))))((((---...(((((((((((...........))))))..)---)))))))).................)).)))))))...... ( -36.50) >consensus UCAACGAGGCUGUGGAGCACCGCCGAC___AAAUCCUCAAAUCACCAAGUGCUAUGAAUGCGAUGAAGGCGCUGGGUGUAAUAGAAAGGAUUUCACCCGGUGCUAUCA .....(((((.((((....)))).).........))))...........(((.......))).....(((((((((((.(((.......))).))))))))))).... (-20.28 = -20.03 + -0.25)

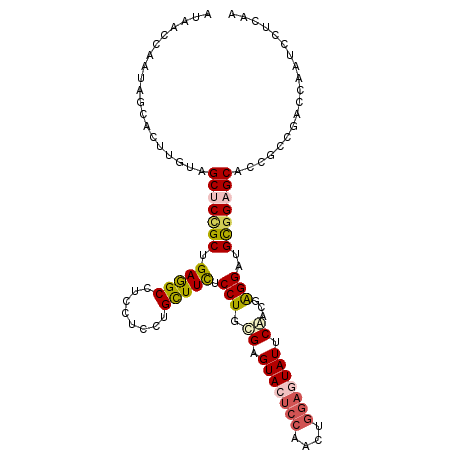
| Location | 18,984,924 – 18,985,032 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18984924 108 - 22407834 AUAGCUAAUAGCACUUGUAGCUCCGCUGAGGCCUCUUCCUGUUUCUCCUGUGAGUACUCCAACUGGAGUAUUCAGCGAGGCUGUGGAGCACCGCCGGAAAAUCCUCAA ...((.....)).......(((((((...((((((.........(....)((((((((((....))))))))))..)))))))))))))......((.....)).... ( -37.80) >DroPse_CAF1 1181 108 - 1 GUAACCAACAGCACGUGUGGCUCUGCCGAGGCCUCCUCCUGCUUCGCCUGCGAGUACUCCGACUGGAAUAUACGCAGGGGAUGCGGUGCCCCACCCACCGAAGCGGGA ..........((.(...((((...)))).)))....(((((((((.((((((.(((.(((....))).))).))))))((.((.((((...))))))))))))))))) ( -42.90) >DroSim_CAF1 1152 108 - 1 AUAGCUAAUAGCACUUGUAGCUCCGCUGAGGCCUCUUCCUGUUUCUCCUGCGAGUACUCCAACUGGAGUAUUCAACGAGGCUGCGGAGCACCGCCGGCAAAUCCUCAA ..........((...((..(((((((...((((((.....((.......))(((((((((....)))))))))...)))))))))))))..))...)).......... ( -40.90) >DroYak_CAF1 1145 108 - 1 AUAGCUAAUAGCACUUGUAGCUCCGCUGAAGCCUCUUCCUGUUUCUCCUGUGAGUACUCAAACUGGAGUAUUCAACGGGGCUGUGGAGCACCGCCGACCAAUCCUCAA ...((.....)).......(((((((.(((((........)))))((((((((((((((......)))))))).))))))..)))))))................... ( -35.10) >DroAna_CAF1 1039 108 - 1 AUUAGCAAUAGCACCUGUGGCUCAGCAGAAGCCUCCUCCUGCUUUUCCUGCGAGUACGACGACUGGAGUAUCCAGCGGGGAUGCGGAGCACCGCCUAGUAAUCCCGGC ....((....)).((((((.(((.((((((((........))))...))))))))))).((.((((.....))))))(((((((((....))))......))))))). ( -33.50) >DroPer_CAF1 1181 108 - 1 GUAACCAACAGCACGUGUGGCUCUGCCGAGGCCUCCUCCUGCUUCGCCUGCGAGUACUCCGACUGGAAUAUACGCAGAGGAUGCGGUGCCCCACCCACCGAAGCGGGA ..........((.(...((((...)))).)))....(((((((((..(((((.(((.(((....))).))).))))).((.((.((((...))))))))))))))))) ( -39.70) >consensus AUAACCAAUAGCACUUGUAGCUCCGCUGAGGCCUCCUCCUGCUUCUCCUGCGAGUACUCCAACUGGAGUAUUCAACGAGGAUGCGGAGCACCGCCGACCAAUCCUCAA ...................(((((((.(((((........))))).(((.((.(((((((....))))))).))...)))..)))))))................... (-22.57 = -22.77 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:57 2006