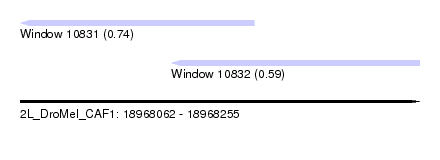
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,968,062 – 18,968,255 |
| Length | 193 |
| Max. P | 0.738686 |

| Location | 18,968,062 – 18,968,175 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -22.89 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
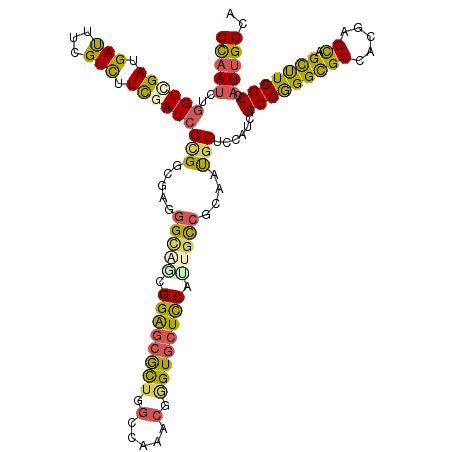
>2L_DroMel_CAF1 18968062 113 - 22407834 GCCCCAUCGCCGGGCGCUACUAGUAGCUUGGCCAUUGCCAAGAUGAUCGCAGACAAGAUCGAGAAUGAGUUCUCUAUCGGCAAGUGAUGCCUCUAUCCCAAAUAAUGU-AACCA-- ((((.......))))((((....))))..(((..(((((.....((((........))))(((((....)))))....))))).....))).................-.....-- ( -31.10) >DroGri_CAF1 1439 104 - 1 GCUCCGUCGCCAGGCGCCACCAGCAGCUUGGCCAUUGCCAAGAUGAUUGCGGACAAGAUCGAGACGGAGUUCUCCAUUGGCAAGGC------------CAAGUAAGAUUAGCUCAA (((..(((((..((.....)).)).((((((((.(((((((..(((((........)))))....(((....))).))))))))))------------)))))..))).))).... ( -36.60) >DroSec_CAF1 650 113 - 1 GCCCCAUCGCCGGGCGCUACUAGUAGCUUGGCCAUUGCAAAGAUGAUCGCAGACAAGAUCGAGAAUGAGUUCUCUAUCGGCAAGUGAUGCCUUUAUCUCAAAUAAUGU-AACCA-- ((((.......))))((((....)))).(((.(((((...((((((..........(((.(((((....))))).)))((((.....)))).))))))....))))).-..)))-- ( -30.00) >DroEre_CAF1 631 113 - 1 GCUCCAUCACCGGGCGCUACUAGUAGCUUGGCCAUUGCCAAGAUGAUCGCAGACAAGAUCGAGACUGAGUUCUCUAUCGGCAAGUGAUGCUAAUGUCCCAAACUUUAU-AAGCA-- (((.((((((..(((((((....))))...))).(((((.....((((........))))((((......))))....))))))))))).(((.((.....)).))).-.))).-- ( -30.60) >DroYak_CAF1 499 113 - 1 GCCCCAUCACCUGGCGCGACUAGCAGCUUGGCCAUUGCCAAGAUGAUCGCAGACAAGAUCGAGACCGAGUUCUCUAUCGGCAAGUGAUGUCAAUAUCCCAAACAAUAU-AAGCA-- ((..((((((.((.(((((....((.((((((....)))))).)).))))......(((.((((......)))).)))).)).))))))...((((........))))-..)).-- ( -32.70) >DroMoj_CAF1 1141 102 - 1 GCGCCAUCGCCGGGCGCAACGAGCAGCUUGGCCAUUGCCAAGAUGAUCGCCGACAAGAUCGAAACGGAGUUUGCCAUUGGCAAGGC------------CAACUAGUUCCAAUUG-- (((((.......)))))...((((((.((((((.(((((((..(((((........)))))....((......)).))))))))))------------))))).))))......-- ( -40.80) >consensus GCCCCAUCGCCGGGCGCUACUAGCAGCUUGGCCAUUGCCAAGAUGAUCGCAGACAAGAUCGAGACUGAGUUCUCUAUCGGCAAGUGAUGCC__UAUCCCAAAUAAUAU_AACCA__ ........(((((((((.....)).)))))))..(((((.....((((........))))((((......))))....)))))................................. (-22.89 = -22.95 + 0.06)
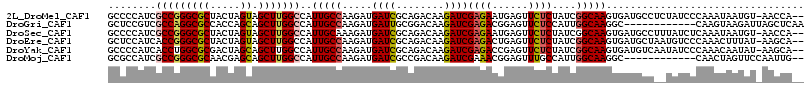
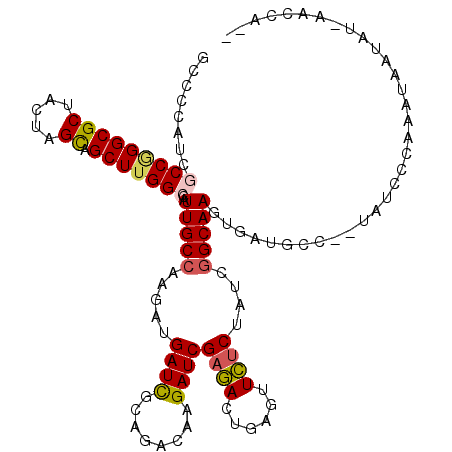
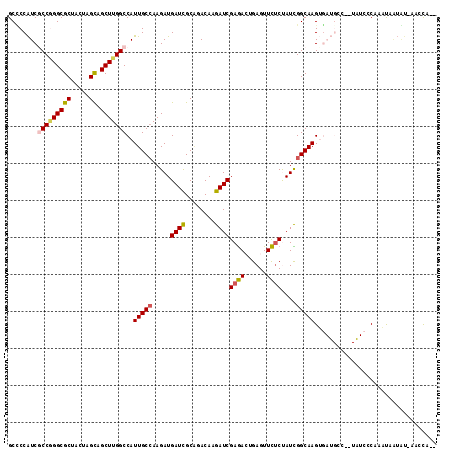
| Location | 18,968,135 – 18,968,255 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -48.75 |
| Consensus MFE | -36.53 |
| Energy contribution | -35.53 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18968135 120 - 22407834 GCAAUCUGGUGGACGAUUUUGUUUUCGAUCGCGGACAAGGAUCGGGAGCGCUAGCCAAGCGAGUGCUUCACUGCAGAAACGCCCCAUCGCCGGGCGCUACUAGUAGCUUGGCCAUUGCCA (((((..(((((.((.((((((...(((((.(......))))))((((((((.((...)).))))))))...)))))).))..)))))(((((((((.....)).))))))).))))).. ( -45.80) >DroVir_CAF1 904 120 - 1 GUAAUCUGGUCGAUGACUUCGUCUUUGAUCGCGGCGAGGGCAGCGGACCGCUGGCCAAACGUGUGCUCCAUUGCCGCAAUGCACCGUCGCCGGGCGCGACGAGCAGCCUGGCCAUUGCCA (..(((.((.(((.....))).))..)))..)(((((((.(((((...))))).))..(((.((((..............))))))).(((((((((.....)).)))))))..))))). ( -49.84) >DroGri_CAF1 1503 120 - 1 GUAAUCUUGUCGAUGAUUUCGUCUUUGAUCGAGGCGAGGGCAGCGGAGCGCUGGCCAAACAGGUGCUCCACUGUCGCAAUGCUCCGUCGCCAGGCGCCACCAGCAGCUUGGCCAUUGCCA .....((((((..(((((........))))).))))))(((((.(((((((((......)).))))))).)))))((((((.......(((((((((.....)).))))))))))))).. ( -51.01) >DroWil_CAF1 956 120 - 1 GCAAUCUGGUUGAUGAUUUCGUCUUCGAUCGCGGCGAGGGUGAGGGAGCAUUGGCGAAACAAGUUCUUCAUUGCCGUAAUGCUCCAUCGCCGGGUGCUACAAGUAGUUUGGCCAUAGCCA ((.(((.((..((((....)))).))))).))(((...(((((.(((((((((((((.............)))))..)))))))).))))).(((((((....))))...)))...))). ( -43.52) >DroMoj_CAF1 1203 120 - 1 GCAAUCUGGUCGAUGAUUUCGUCUUUGAUCGUGGCGAGGGCAGCGGGGCGCUGGCCAAGCGGGUGCUCCAUUGCCGCAACGCGCCAUCGCCGGGCGCAACGAGCAGCUUGGCCAUUGCCA (((((..((((((.(((...))).))))))((((((.((((((.((((((((.((...)).)))))))).))))).)....)))))).(((((((((.....)).))))))).))))).. ( -55.60) >DroAna_CAF1 739 120 - 1 GCAACCUGGUGGAUGACUUUGUCUUCGAUCGCGGAGAGGGUUCGGGAGCACUGGCCAAGAGGGUGCUUCAUUGCCGAAAUGCUCCGUCGCCAGGUGCCACGAGCAGCUUGGCCAUUGCAA (((((((((((((.(((...))).))....((((((....((((((((((((..(...)..))))))).....)))))...))))))))))))))((((.(.....).))))...))).. ( -46.70) >consensus GCAAUCUGGUCGAUGAUUUCGUCUUCGAUCGCGGCGAGGGCAGCGGAGCGCUGGCCAAACGGGUGCUCCAUUGCCGCAAUGCUCCAUCGCCGGGCGCCACGAGCAGCUUGGCCAUUGCCA (((((..((((((.(((...))).))))))(((.....(((((.((((((((.(.....).)))))))).)))))....)))......(((((((((.....)).))))))).))))).. (-36.53 = -35.53 + -0.99)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:53 2006