| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,952,333 – 18,952,434 |
| Length | 101 |
| Max. P | 0.598530 |

| Location | 18,952,333 – 18,952,434 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.14 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -10.23 |
| Energy contribution | -9.82 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18952333 101 - 22407834 GAGGCAGAGGCAGUCACCGAAUUAGUACUGGACAACUUUCCGGAUGCCAAAUUCUGGGUCUCCCUGCAGUGCAUGGUGAGUAA-----UA-UCGCAAA-AAACAAUAA ...((((.((.((.(.(((((((.((((((((......))))).)))..))))).))).)))))))).((.....((((....-----..-))))...-..))..... ( -28.30) >DroPse_CAF1 6249 93 - 1 GAGGCACUCGCCAUCACUGAAAGCAUACUCGAGAACUAUACGAAUGUCAAGUUUUGGGUAUCCUUUCAGUGCAAGGUGAGUUG-----UG-CU---------UUGGAU ((((((((((((..(((((((((.((((((.((((((..((....))..)))))))))))).)))))))))...)))))...)-----))-))---------)).... ( -37.40) >DroSec_CAF1 7363 102 - 1 GAAGCAGAGGCAGUCACCGAAUUAGUACUGGACAACUUUCCGGAUGCCAAAUUCUGGGUCUCCCUGCAGUGCAUGGUCAGUAA-----UA-UCGGAAAAAAAAUGGCG ...((((.((.((.(.(((((((.((((((((......))))).)))..))))).))).))))))))........((((....-----..-............)))). ( -25.17) >DroSim_CAF1 6926 101 - 1 GAAGCAGAGGCAGUCACCGAAUUAGUACUGGACAACUUUCCGGAUGCCAAAUUCUGGGUCUCCCUGCAGUGCAUGGUGAGUAA-----UA-UAGGAAA-AAAAUGGAG ...((((.((.((.(.(((((((.((((((((......))))).)))..))))).))).))))))))................-----..-.......-......... ( -23.40) >DroAna_CAF1 9436 104 - 1 GAGGCAGAGGCUGUUACCGAACUUGUACUGGAUUCCUAUGAGGGUGUCAAUUUCUGGGUGUCCUUCCAGUGUCAGGUGAGUGUGGAAGACAUCAACUG----UUGAAG ..(((((..(.((((.(((.((((((((((((........((((..((........))..))))))))))).))))).....)))..)))).)..)))----)).... ( -29.90) >DroPer_CAF1 6242 93 - 1 GAGGCACUCGCCAUCACUGAAAGCAUACUCGAGAACUAUACGAAUGUCAAGUUUUGGGUAUCCUUUCAGUGCAAGGUGAGUUG-----UG-CU---------UUGGAU ((((((((((((..(((((((((.((((((.((((((..((....))..)))))))))))).)))))))))...)))))...)-----))-))---------)).... ( -37.40) >consensus GAGGCAGAGGCAGUCACCGAAUUAGUACUGGACAACUAUCCGGAUGCCAAAUUCUGGGUCUCCCUGCAGUGCAAGGUGAGUAA_____UA_UCG_AAA____UUGGAG ...((....))..(((((......((((((...........((((.....)))).(((...)))..))))))..)))))............................. (-10.23 = -9.82 + -0.41)


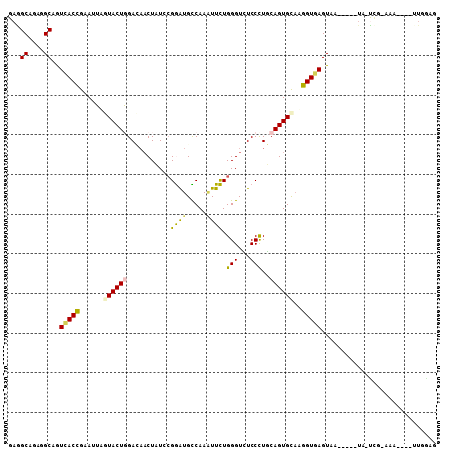
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:46 2006