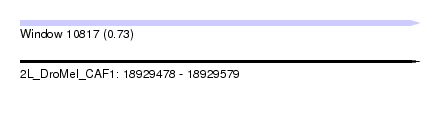
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,929,478 – 18,929,579 |
| Length | 101 |
| Max. P | 0.732189 |

| Location | 18,929,478 – 18,929,579 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.73 |
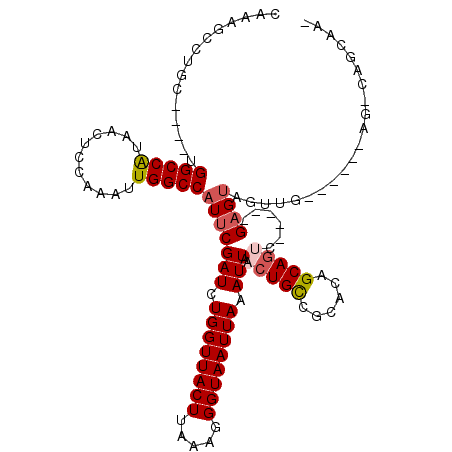
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -18.36 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18929478 101 + 22407834 CAAGGCCUGC----CGGCCAUAACUCCAAAUUGGCCAUUCGAUCUGGUUACUUAAAGGGUAAUUAAAUUAACUGCCGCACAGCAGUC-----GAGUAGUUG------AGCCAGCAA- ...((((.((----.(((((...........)))))((((((..((((((((.....))))))))......((((......))))))-----)))).)).)------.))).....- ( -29.00) >DroPse_CAF1 56197 116 + 1 CAAAGUCUUCUACUUGGCCAUAACUCCAAAUUGGCCAUUCGAUAUGGUUACUUAAAGGGUAAUUAAAUUAACUGCCGCACAGCAGUGGCCCAGAGUACGAAAACACAGG-CAGUAAA ....((.((((((((((((...((((..((.(((((((.....))))))).))...))))..........(((((......)))))))))..))))).))).)).....-....... ( -28.80) >DroEre_CAF1 53413 100 + 1 CAAGGCCUGC----UGGCCAUAACUCCAAAUUGGCCAUUCGAUCUGGUUACUUAAAAGGUAAUUAAAUUAACUGCCGCACAGCAGUC-----GAGUAGUUG------AG-CAGCGA- ....((.(((----((((((...........)))))((((((..((((((((.....))))))))......((((......))))))-----)))).....------))-))))..- ( -29.80) >DroYak_CAF1 53804 100 + 1 CAAGGCCUGC----UGGCCGAAUCUCCAAAUUGGCCAUUCGAUCUGGUUACUUAAAGGGUAAUUAAAUUAUCUGCCGCACAGCAGUC-----GUGUAGUUG------AG-CAACGA- ......((((----(((((((((..((.....))..)))))...((((((((.....))))))))...........)).))))))((-----((((.....------.)-).))))- ( -25.30) >DroAna_CAF1 37238 99 + 1 CCAAGUAUGU----CGGCCAUAACUCCAAAUUGGCCAUUCGAUCUGGUUACUUAAAGGGUAAUUAAAUUAAAUGUCGCACAGCAGCUCCCAGGAGCAGUGG------CG-------- ..........----.(((((...........)))))(((..((.((((((((.....)))))))).))..)))..(((...((.((((....)))).)).)------))-------- ( -26.70) >DroPer_CAF1 58296 116 + 1 CAAAGUCUUCUACUUGGCCAUAACUCCAAAUUGGCCAUUCGAUAUGGUUACUUAAAGGGUAAUUAAAUUAACUGCCGCACAGCAGUGGCCCAGAGUACGAAAACACAGG-CAGUAAA ....((.((((((((((((...((((..((.(((((((.....))))))).))...))))..........(((((......)))))))))..))))).))).)).....-....... ( -28.80) >consensus CAAAGCCUGC____UGGCCAUAACUCCAAAUUGGCCAUUCGAUCUGGUUACUUAAAGGGUAAUUAAAUUAACUGCCGCACAGCAGUC_____GAGUAGUUG______AG_CAGCAA_ ...............(((((...........)))))(((((((.((((((((.....)))))))).))).(((((......)))))......))))..................... (-18.36 = -18.92 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:39 2006