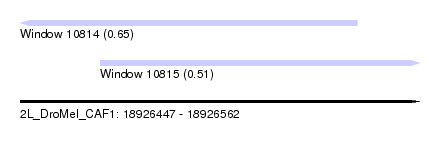
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,926,447 – 18,926,562 |
| Length | 115 |
| Max. P | 0.649905 |

| Location | 18,926,447 – 18,926,544 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -28.49 |
| Consensus MFE | -19.60 |
| Energy contribution | -19.05 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18926447 97 - 22407834 ------GGACCAAAAACUGGUGCAAAUUAACACGCUAAGCACUUGAAUCGGACCAAAUGUGGCAUGAGUAAUGCCCUCAAGUGG-----------------CCAACAGAUGGCGCACAUU ------(.((((.....)))).)..........((....(((((((.....((.....))(((((.....))))).)))))))(-----------------(((.....))))))..... ( -26.30) >DroVir_CAF1 46230 101 - 1 UUCGAAGCACUGA-AACUGGUGCAAAUUAACACGCUAAGCACUUGAAU-GCGCCAAAUGUGGCGUGAGUAAUGCCCUCAAGUGG-----------------CCGACAGAUGGCGCACAUU (((((.(((((..-....)))))..........((...))..)))))(-((((((..((((((.((((.......))))....)-----------------)).)))..))))))).... ( -32.20) >DroPse_CAF1 53148 113 - 1 ------GAUCUAG-AACUGGUGCAAAUUAACACGCUAAGCACUUGAAUCCGGCCAAAUGUGGCAUGAGUAAUGCCCUCAAGUGGCUUCGCCUCGCAUCGUUCCAACAGAUGGCGCACAUU ------.......-.....((((..........((.((((((((((.....((.....))(((((.....))))).)))))).)))).))...(((((.........))).))))))... ( -28.20) >DroGri_CAF1 69534 89 - 1 ------------A-AACUGGUGCAAAUUAACGCGCUAAGCACUUGAAU-GCGCCAAAUGUGGCGUGAGUAAUGCCCUCAAGUGG-----------------CCGACAGAUGGCGCACAUU ------------.-...((((((........))))))..........(-((((((..((((((.((((.......))))....)-----------------)).)))..))))))).... ( -30.30) >DroMoj_CAF1 74395 89 - 1 ------------A-AACUGGUGCAAAUUAACACGCUAAGCACUUGAAC-GCGCUAAAUGUGGCGUGAGUAAUGCCUCCAAGUGG-----------------ACAACAGAUGGCGUGCAUU ------------.-..(.(((((...............))))).)..(-((((((..((((((((.....)))))(((....))-----------------)..)))..))))))).... ( -25.76) >DroPer_CAF1 55229 113 - 1 ------GAUCUAG-AACUGGUGCAAAUUAACACGCUAAGCACUUGAAUCCGGCCAAAUGUGGCAUGAGUAAUGCCCUCAAGUGGCUUCGCCUCGCAUCGUUCCAACAGAUGGCGCACAUU ------.......-.....((((..........((.((((((((((.....((.....))(((((.....))))).)))))).)))).))...(((((.........))).))))))... ( -28.20) >consensus ______G__CUAA_AACUGGUGCAAAUUAACACGCUAAGCACUUGAAU_GCGCCAAAUGUGGCAUGAGUAAUGCCCUCAAGUGG_________________CCAACAGAUGGCGCACAUU ...................((((................(((((((.....((.....))(((((.....))))).)))))))..................(((.....))).))))... (-19.60 = -19.05 + -0.55)

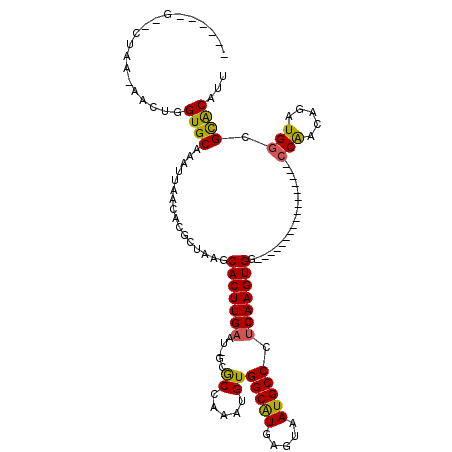
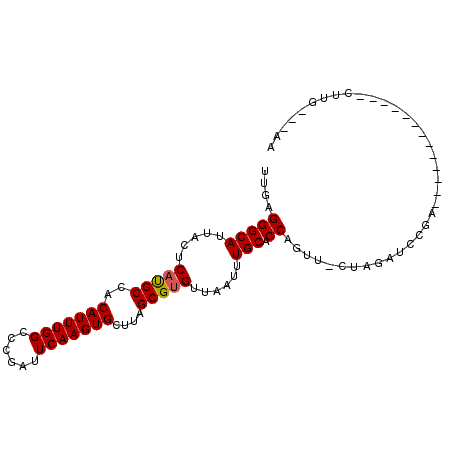
| Location | 18,926,470 – 18,926,562 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.53 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18926470 92 + 22407834 UUGAGGGCAUUACUCAUGCCACAUUUGGUCCGAUUCAAGUGCUUAGCGUGUUAAUUUGCACCAGUUUUUGGUCCCAGUAUCGG-GA---UCUUU---AA ....(((((.....(((((..(((((((......)))))))....)))))......))).)).......((((((......))-))---))...---.. ( -26.60) >DroPse_CAF1 53188 82 + 1 UUGAGGGCAUUACUCAUGCCACAUUUGGCCGGAUUCAAGUGCUUAGCGUGUUAAUUUGCACCAGUU-CUAGAUCCGA-------------CUUG---AG (..(((((((.....))))).........((((((..((.(((..(.((((......)))))))).-)).)))))).-------------))..---). ( -20.90) >DroGri_CAF1 69557 75 + 1 UUGAGGGCAUUACUCACGCCACAUUUGGCGC-AUUCAAGUGCUUAGCGCGUUAAUUUGCACCAGUU-U------CGA-------------CUCG---AA ((((((((.........)))....((((.((-(.....((((...)))).......))).))))..-.------...-------------))))---). ( -19.40) >DroWil_CAF1 18795 91 + 1 UUGAGGGCAUUACUCACGCCACAUUUGGCGC-AUUCAAGUGCUUAGCGUGUUAAUUUGCACCAGUU-UCAGUUG--GUCUUGG-GUCGUCGUUG---AA ..((.(((((((..(((((.......(((((-......)))))..))))).)))).....((((..-((....)--)..))))-))).))....---.. ( -22.10) >DroAna_CAF1 34465 95 + 1 UUGAGGGCAUUACUCAUGCCACAUUUGGCAGGAUUCAAGUGCUUAGCGUGUUAAUUUGCACCAGUU-CCUGCCGCAGCUCCUGAGC---CCUUUUCGAG ..((((((..................(((((((.....((((...............))))....)-)))))).(((...))).))---))))...... ( -28.46) >DroPer_CAF1 55269 82 + 1 UUGAGGGCAUUACUCAUGCCACAUUUGGCCGGAUUCAAGUGCUUAGCGUGUUAAUUUGCACCAGUU-CUAGAUCCGA-------------CUUG---AG (..(((((((.....))))).........((((((..((.(((..(.((((......)))))))).-)).)))))).-------------))..---). ( -20.90) >consensus UUGAGGGCAUUACUCAUGCCACAUUUGGCCCGAUUCAAGUGCUUAGCGUGUUAAUUUGCACCAGUU_CUAGAUCCGA_____________CUUG___AA ....(((((.....(((((..(((((((......)))))))....)))))......))).))..................................... (-16.59 = -16.53 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:38 2006