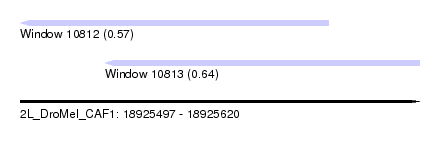
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,925,497 – 18,925,620 |
| Length | 123 |
| Max. P | 0.643433 |

| Location | 18,925,497 – 18,925,592 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -20.39 |
| Energy contribution | -19.58 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.571014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18925497 95 - 22407834 UAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUGUU-------CGGGGUCCUUCUAUGGCUCUGGGACGGAUCUUA--AGGAG-- ................(((((.......((((((....))))))....(((((.(((-------(.((..((......))..)).)))))))))...--)))))-- ( -30.90) >DroPse_CAF1 51872 87 - 1 UAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCGCCUCACGGUCCUGUU-------CAGGGUCU----------CGGUGCGGGAUUUUC--AGGAGUU .......((((((...............((((((....))))))....(..(((...-------.)))..).----------.))))))........--....... ( -23.90) >DroEre_CAF1 49168 95 - 1 UAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUGUU-------CAGGGUCCUGCUAUGGCUCUGGGACGGAUCUUA--AGAAG-- ................(((((.......((((((....))))))....(((((..((-------(((((.((......)))))))))..)))))...--)))))-- ( -31.80) >DroYak_CAF1 49756 95 - 1 UAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUGUU-------CGGGGUCCUUCUCCGGCUCUGGGACGGAUCUUA--AGAAG-- ................(((((.......((((((....))))))....(((((.(((-------(.((..((......))..)).)))))))))...--)))))-- ( -30.80) >DroAna_CAF1 33697 97 - 1 UAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUGCUCCAGCUCCAGGGUCCUUCUAUAGCCCC-------AUCGUGGCAGGGG-- .......(((..................((((((....)))))).((((((...((....))....((((.........)))).-------)))))))))....-- ( -24.90) >DroPer_CAF1 53972 87 - 1 UAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCGCCUCACGGUCCUGUU-------CAGGGUCU----------CGGUGCGGGAUUUUC--AGGAGUU .......((((((...............((((((....))))))....(..(((...-------.)))..).----------.))))))........--....... ( -23.90) >consensus UAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUGUU_______CAGGGUCCUUCUAUGGCUCUGGGACGGAUCUUA__AGGAG__ ................(((((.......((((((....))))))....(..((((.........))))..)............................))))).. (-20.39 = -19.58 + -0.80)

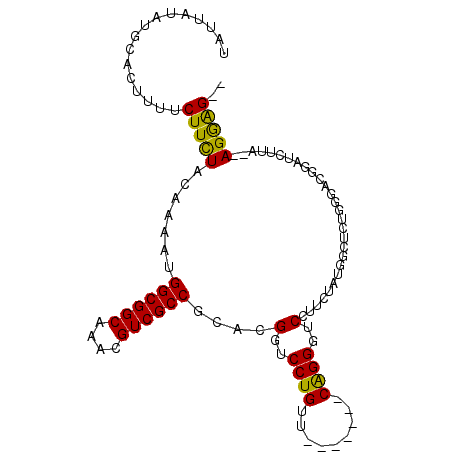
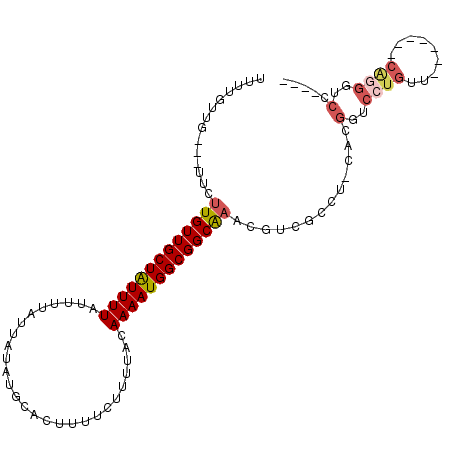
| Location | 18,925,523 – 18,925,620 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -20.11 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18925523 97 - 22407834 UUUUGUUG---GCCGUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCG-CACGGUCCUGUU-------CGGGGUCCUUCU ........---(((((((((((((..((...))..))).))).................((((((....)))))))-))))))(((...-------.)))........ ( -24.10) >DroVir_CAF1 45202 87 - 1 UUUUGCUGUUUUUGUUGUUGCUGUUUUAUAUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCGAACGUCGCCAACAUC---CUGAU-------C----------- .........((((((.(....(((..((((....)))).))).....)....))))))(((((((....))))))).....---.....-------.----------- ( -15.10) >DroPse_CAF1 51894 93 - 1 UUUUGUUG---UUCUUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCGCCU-CACGGUCCUGUU-------CAGGGUCU---- ((((((..---.......((((((..((...))..))).)))..........)))))).((((((....)))))).-...(..(((...-------.)))..).---- ( -20.01) >DroWil_CAF1 17756 89 - 1 UCGCAUUG------UUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCC--U-CCUGG---UGUU-------GGGGACCCUUCU ........------(((((((((((((...........................)))))))))))))..((((--.-((...---....-------))))))...... ( -18.83) >DroAna_CAF1 33718 97 - 1 UU----------CCUUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCG-CACGGUCCUGCUCCAGCUCCAGGGUCCUUCU ..----------(((((..(((.....................................((((((....))))))(-((......)))...)))..)))))....... ( -22.60) >DroPer_CAF1 53994 93 - 1 UUUUGUUG---UUCUUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCGCCU-CACGGUCCUGUU-------CAGGGUCU---- ((((((..---.......((((((..((...))..))).)))..........)))))).((((((....)))))).-...(..(((...-------.)))..).---- ( -20.01) >consensus UUUUGUUG___UUCUUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCGCCU_CACGGUCCUGUU_______CAGGGUCC____ ..............(((((((((((((...........................))))))))))))).............(..((((.........))))..)..... (-11.13 = -11.90 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:36 2006