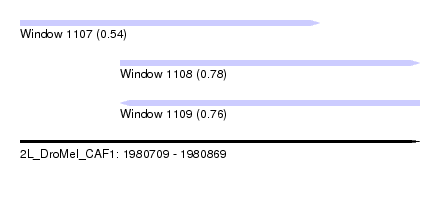
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,980,709 – 1,980,869 |
| Length | 160 |
| Max. P | 0.779085 |

| Location | 1,980,709 – 1,980,829 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -23.56 |
| Energy contribution | -22.88 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1980709 120 + 22407834 AAAAAUACGGAUAAAUGGUUUGCUAUUCCUAAUAAAAUAAAGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCCGCUGCGGUCACACUGCGGCCAGGAGGAAAUAAAGGUAUCAAA .........((((....((((.(...((((..................(..((((((....))))))..)....((((((.(.(....).).))))))))))).)))).....))))... ( -29.50) >DroSim_CAF1 6683 120 + 1 AAAAAUACGGAUAAAUGGUUUGCUAUUCCUAAUAAAAUAAAGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCCGCUGCGGUCACACUGCGGCCAGGAGGAAAUAAAGGUAUCAAA .........((((....((((.(...((((..................(..((((((....))))))..)....((((((.(.(....).).))))))))))).)))).....))))... ( -29.50) >DroEre_CAF1 6478 120 + 1 AAAAAUACGGAUAAAUGGUUUGCUAUUCCUAAUAAAAUAAAGCAAUAAGAAUAUCGAACUAUCGAUGAGCGAUAGGCUCGUGCGGUCACACUACGGCUAGGAGGAAAUAAAGGUAUCGAA .........((((....((((.(...((((...........(((........(((((....)))))((((.....)))).)))((((.......))))))))).)))).....))))... ( -23.00) >DroYak_CAF1 6481 120 + 1 AAAAAUACGGAUAAAUGGUUUGCUAUUCCUAAUAAAAUAAAGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCUCCUGCGGUCACACUACGGCUAGGAGGAAAUAAAGGUAUCGAA ........(((...((((....)))))))........................((((.((((((.....)))))).(((((((.((......)).)).)))))............)))). ( -25.70) >consensus AAAAAUACGGAUAAAUGGUUUGCUAUUCCUAAUAAAAUAAAGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCCCCUGCGGUCACACUACGGCCAGGAGGAAAUAAAGGUAUCAAA .........((((....((((.(...((((...........(((.......((((((....))))))(((.....)))..)))((((.......))))))))).)))).....))))... (-23.56 = -22.88 + -0.69)

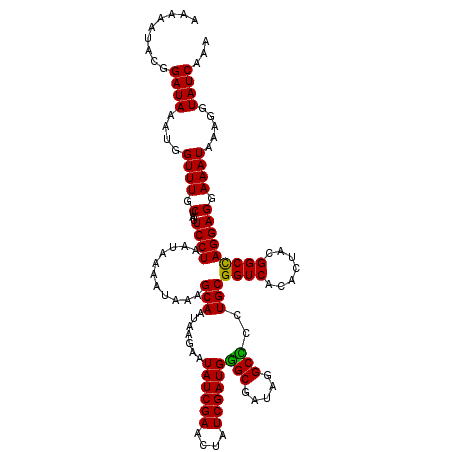
| Location | 1,980,749 – 1,980,869 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -26.44 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1980749 120 + 22407834 AGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCCGCUGCGGUCACACUGCGGCCAGGAGGAAAUAAAGGUAUCAAACUAUCGCAGGUGGCAAGGCAGCGAUAGAUCGGAUAGAAAA .............((((.(((((....(((.....)))(((((.(((((.((((((..((...((.((....)).))...)).)))))))))))...)))))))))).))))........ ( -41.40) >DroSim_CAF1 6723 120 + 1 AGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCCGCUGCGGUCACACUGCGGCCAGGAGGAAAUAAAGGUAUCAAACUAUCGCAGGUGGCAAGGCAGCGAUAGAUCGGAUGGAAAA .............((((.(((((....(((.....)))(((((.(((((.((((((..((...((.((....)).))...)).)))))))))))...)))))))))).))))........ ( -41.40) >DroEre_CAF1 6518 115 + 1 AGCAAUAAGAAUAUCGAACUAUCGAUGAGCGAUAGGCUCGUGCGGUCACACUACGGCUAGGAGGAAAUAAAGGUAUCGAACUAUCGCAGGUGGUAGGCCAGUGAUAUAUCGA-----AUA ...........((((...((((((.....)))))).(((.(((.((......)).)).).)))........))))((((..((((((.(((.....))).))))))..))))-----... ( -28.40) >DroYak_CAF1 6521 120 + 1 AGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCUCCUGCGGUCACACUACGGCUAGGAGGAAAUAAAGGUAUCGAACUAUCGCAGGUGGUAGGCCUGCGAUAGAUCGGAUGGGAUA ...........((((...((((((.....)))))).(((((((.((......)).)).)))))........))))((((.(((((((((((.....))))))))))).))))........ ( -43.20) >consensus AGCAAUAAGAAUAUCGAACUAUCGAUGGGCGAUAGGCCCCUGCGGUCACACUACGGCCAGGAGGAAAUAAAGGUAUCAAACUAUCGCAGGUGGCAAGCCAGCGAUAGAUCGGAUGGAAAA .............((((.((((((...(((.....)))(((...(((((.((((((.(((...((.((....)).))...))))))))))))))..)))..)))))).))))........ (-26.44 = -27.12 + 0.69)

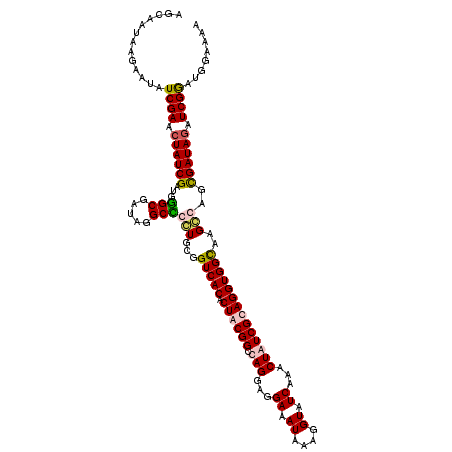
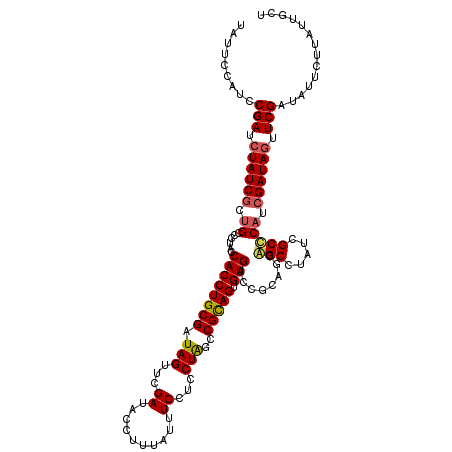
| Location | 1,980,749 – 1,980,869 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1980749 120 - 22407834 UUUUCUAUCCGAUCUAUCGCUGCCUUGCCACCUGCGAUAGUUUGAUACCUUUAUUUCCUCCUGGCCGCAGUGUGACCGCAGCGGCCUAUCGCCCAUCGAUAGUUCGAUAUUCUUAUUGCU .........(((.((((((((((...(.((((((((.(((...((..........))...)))..))))).))).).)))))(((.....)))....))))).))).............. ( -33.00) >DroSim_CAF1 6723 120 - 1 UUUUCCAUCCGAUCUAUCGCUGCCUUGCCACCUGCGAUAGUUUGAUACCUUUAUUUCCUCCUGGCCGCAGUGUGACCGCAGCGGCCUAUCGCCCAUCGAUAGUUCGAUAUUCUUAUUGCU .........(((.((((((((((...(.((((((((.(((...((..........))...)))..))))).))).).)))))(((.....)))....))))).))).............. ( -33.00) >DroEre_CAF1 6518 115 - 1 UAU-----UCGAUAUAUCACUGGCCUACCACCUGCGAUAGUUCGAUACCUUUAUUUCCUCCUAGCCGUAGUGUGACCGCACGAGCCUAUCGCUCAUCGAUAGUUCGAUAUUCUUAUUGCU ...-----((((..((((.(.((.......)).).))))..)))).................(((.(((((((....))))((((.....))))(((((....))))).....))).))) ( -24.70) >DroYak_CAF1 6521 120 - 1 UAUCCCAUCCGAUCUAUCGCAGGCCUACCACCUGCGAUAGUUCGAUACCUUUAUUUCCUCCUAGCCGUAGUGUGACCGCAGGAGCCUAUCGCCCAUCGAUAGUUCGAUAUUCUUAUUGCU .........(((.((((((((((.......)))))))))).))).............(((((.((.((......)).))))))).((((((.....))))))..(((((....))))).. ( -36.10) >consensus UAUUCCAUCCGAUCUAUCGCUGCCCUACCACCUGCGAUAGUUCGAUACCUUUAUUUCCUCCUAGCCGCAGUGUGACCGCAGCAGCCUAUCGCCCAUCGAUAGUUCGAUAUUCUUAUUGCU .........(((.((((((.((......((((((((.(((...((..........))...)))..))))).)))........(((.....))))).)))))).))).............. (-22.08 = -22.32 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:40 2006