| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,919,152 – 18,919,258 |
| Length | 106 |
| Max. P | 0.956045 |

| Location | 18,919,152 – 18,919,258 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -30.68 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
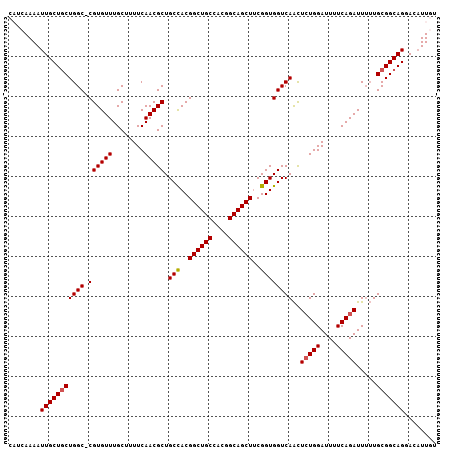
>2L_DroMel_CAF1 18919152 106 + 22407834 CAUCAAAAUUGCUGCUGGCCCGUGUUUGCUUUUCAACGCUGCCACGGCUGCCACGGCAGCUUCGGUGGUCAAUUCUGGAUUUUCAGAUU-UUGCGGCAGGACAUUGU ........((((((((((((.(((((........))))).(((..((((((....))))))..))))))))..(((((....)))))..-..)))))))........ ( -36.50) >DroSec_CAF1 44241 106 + 1 CAUCAAAAUUGCUGCUGGC-CGUGUUUGCGUUUCAACGCUGCCACGGCUGCCACGGCAGCUUCGGUGGUCAACUCUGGAUUUUCAGAUUUUUGCGGCAGGACAUUGU ........(((((((((((-(......((((....)))).(((..((((((....))))))..))))))))..(((((....))))).....)))))))........ ( -38.20) >DroSim_CAF1 44321 106 + 1 CAUCAAAAUUGCUGCUGGC-CGUGUUUGCGUUUCAACGCUGCCACGGCUGCCACGGCAGCUUCGGUGGUCAACUCUGGAUUUUCAGAUUUUUGCGGCAGGACAUUGU ........(((((((((((-(......((((....)))).(((..((((((....))))))..))))))))..(((((....))))).....)))))))........ ( -38.20) >DroEre_CAF1 42855 102 + 1 ----AAAAUUGCUGCUGGC-CGUGUUUGCUUUUCAACGCUGCCACGGCUGCCACGGCAGCUUCGGUGGUCAACUGUGGAUUUUCAGAUUUUUGCGGCAGGACAUUGU ----....(((((((((((-((((((........))))).(((..((((((....))))))..)))))))).(((........)))......)))))))........ ( -36.00) >DroYak_CAF1 43317 102 + 1 ----AAAAUUGCUUCUGGC-CGUGUUUGCUUUUCAACGCUGCUACGGCUGCCACUGCAGCUCCGGUGGUCAGCUCUGGAUUUUCAGAUUUAUGCGGCAGGACUUUGU ----.........(((.((-((((((((...((((..((((((((((((((....))))))...)))).))))..))))....)))))....))))).)))...... ( -33.40) >consensus CAUCAAAAUUGCUGCUGGC_CGUGUUUGCUUUUCAACGCUGCCACGGCUGCCACGGCAGCUUCGGUGGUCAACUCUGGAUUUUCAGAUUUUUGCGGCAGGACAUUGU ........(((((((((((.((((((........))))).(((..((((((....))))))..))))))))..(((((....))))).....)))))))........ (-30.68 = -30.92 + 0.24)

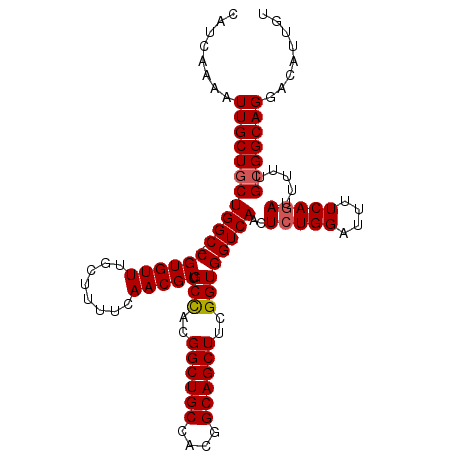
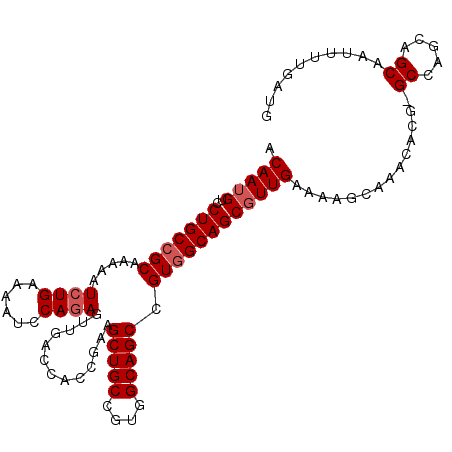
| Location | 18,919,152 – 18,919,258 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18919152 106 - 22407834 ACAAUGUCCUGCCGCAA-AAUCUGAAAAUCCAGAAUUGACCACCGAAGCUGCCGUGGCAGCCGUGGCAGCGUUGAAAAGCAAACACGGGCCAGCAGCAAUUUUGAUG .(((....((((.((..-..((((......))))........(((..((((((((((...))))))))))(((....))).....)))))..)))).....)))... ( -33.10) >DroSec_CAF1 44241 106 - 1 ACAAUGUCCUGCCGCAAAAAUCUGAAAAUCCAGAGUUGACCACCGAAGCUGCCGUGGCAGCCGUGGCAGCGUUGAAACGCAAACACG-GCCAGCAGCAAUUUUGAUG .............((.....((((......))))((((.((.(((..(((((....)))))..)))..((((....))))......)-).)))).)).......... ( -31.20) >DroSim_CAF1 44321 106 - 1 ACAAUGUCCUGCCGCAAAAAUCUGAAAAUCCAGAGUUGACCACCGAAGCUGCCGUGGCAGCCGUGGCAGCGUUGAAACGCAAACACG-GCCAGCAGCAAUUUUGAUG .............((.....((((......))))((((.((.(((..(((((....)))))..)))..((((....))))......)-).)))).)).......... ( -31.20) >DroEre_CAF1 42855 102 - 1 ACAAUGUCCUGCCGCAAAAAUCUGAAAAUCCACAGUUGACCACCGAAGCUGCCGUGGCAGCCGUGGCAGCGUUGAAAAGCAAACACG-GCCAGCAGCAAUUUU---- ....(((.((((((....................((((.((((....(((((....))))).))))))))(((....))).....))-).)))..))).....---- ( -29.70) >DroYak_CAF1 43317 102 - 1 ACAAAGUCCUGCCGCAUAAAUCUGAAAAUCCAGAGCUGACCACCGGAGCUGCAGUGGCAGCCGUAGCAGCGUUGAAAAGCAAACACG-GCCAGAAGCAAUUUU---- .....(((((((.((.....((((......)))).(((.....))).)).)))).))).(((((....((........))....)))-)).............---- ( -26.60) >consensus ACAAUGUCCUGCCGCAAAAAUCUGAAAAUCCAGAGUUGACCACCGAAGCUGCCGUGGCAGCCGUGGCAGCGUUGAAAAGCAAACACG_GCCAGCAGCAAUUUUGAUG .(((((..(((((((.....((((......)))).............(((((....))))).))))))))))))..............((.....)).......... (-26.60 = -27.20 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:33 2006