| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,916,732 – 18,916,831 |
| Length | 99 |
| Max. P | 0.761824 |

| Location | 18,916,732 – 18,916,831 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.27 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -20.19 |
| Energy contribution | -20.07 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18916732 99 + 22407834 CCCACUUCCUACCCCUUUCAACUGGCUACCUUUUGGGCCAGGACACUCCACACUUGUUAAUCGCCCGGAGCGUUCAAAGGCACGGGGUAGAACACGACU ........(((((((......((((((........))))))(((.((((.................)))).))).........)))))))......... ( -29.73) >DroSec_CAF1 41859 92 + 1 CCCACUUC----CC-CUUCAACUGGCUACCUUUUGGGCCAGGACACUCCACACUUGUUAAUCGCCCGGAGCGUUCAAAGGCACGGGGUGGAA--CGACU .(((((((----.(-(((...((((((........))))))(((.((((.................)))).)))..))))...)))))))..--..... ( -27.93) >DroSim_CAF1 41458 96 + 1 CCCACUUCCUACCC-CUUCAACUGGCUACCUUUUGGGCCAGGACACUCCACACUUGUUAAUCGCCCGGAGCGUUCAAAGGCACGGGGUAGAA--CGACU ........((((((-(.....((((((........))))))(((.((((.................)))).))).........)))))))..--..... ( -29.73) >DroEre_CAF1 40380 96 + 1 CCCACAUCCCAGCC-CUUCAACUGGCCACCUUUUGGGCCAGGCCACUCCACACUUGUUAAUCGCCCGGAGCGUUCAAAGGCACGGGGCAAAA--CAACU ...........(((-((....((((((........))))))(((.....((....))....(((.....)))......)))..)))))....--..... ( -27.00) >DroYak_CAF1 40860 90 + 1 CCCACGUCCUACCC-AUUCAAAUGGCUACCUUUCGGGCCAGGAUACUCCACACUUGUUAAUCGCCCGGAGCGUUC------ACGGGGCAAAA--CAACU .....(((((..((-((....))))..((.((((((((..((.....))((....)).....)))))))).))..------..)))))....--..... ( -22.40) >consensus CCCACUUCCUACCC_CUUCAACUGGCUACCUUUUGGGCCAGGACACUCCACACUUGUUAAUCGCCCGGAGCGUUCAAAGGCACGGGGUAGAA__CGACU (((...................(((((........)))))((((.((((.................)))).))))........)))............. (-20.19 = -20.07 + -0.12)

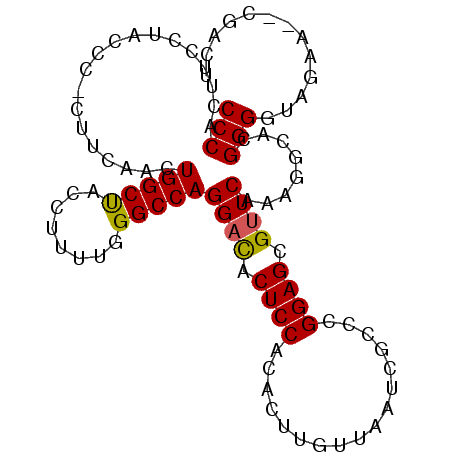
| Location | 18,916,732 – 18,916,831 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.27 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.82 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18916732 99 - 22407834 AGUCGUGUUCUACCCCGUGCCUUUGAACGCUCCGGGCGAUUAACAAGUGUGGAGUGUCCUGGCCCAAAAGGUAGCCAGUUGAAAGGGGUAGGAAGUGGG ..(((..(((((((((..........((((((((.((.........)).)))))))).(((((((....))..)))))......)))))))))..))). ( -40.20) >DroSec_CAF1 41859 92 - 1 AGUCG--UUCCACCCCGUGCCUUUGAACGCUCCGGGCGAUUAACAAGUGUGGAGUGUCCUGGCCCAAAAGGUAGCCAGUUGAAG-GG----GAAGUGGG .....--..((((....(.((((..(((((((((.((.........)).)))))))).(((((((....))..))))))..)))-).----)..)))). ( -34.60) >DroSim_CAF1 41458 96 - 1 AGUCG--UUCUACCCCGUGCCUUUGAACGCUCCGGGCGAUUAACAAGUGUGGAGUGUCCUGGCCCAAAAGGUAGCCAGUUGAAG-GGGUAGGAAGUGGG .....--(((((((((..........((((((((.((.........)).)))))))).(((((((....))..))))).....)-))))))))...... ( -38.20) >DroEre_CAF1 40380 96 - 1 AGUUG--UUUUGCCCCGUGCCUUUGAACGCUCCGGGCGAUUAACAAGUGUGGAGUGGCCUGGCCCAAAAGGUGGCCAGUUGAAG-GGCUGGGAUGUGGG .....--..(..(((((.((((((.(((((((((.((.........)).))))))((((..(((.....))))))).))).)))-)))))))..)..). ( -39.50) >DroYak_CAF1 40860 90 - 1 AGUUG--UUUUGCCCCGU------GAACGCUCCGGGCGAUUAACAAGUGUGGAGUAUCCUGGCCCGAAAGGUAGCCAUUUGAAU-GGGUAGGACGUGGG ....(--((((((((...------....((((((.((.........)).))))))....((((((....))..)))).......-)))))))))..... ( -32.60) >consensus AGUCG__UUCUACCCCGUGCCUUUGAACGCUCCGGGCGAUUAACAAGUGUGGAGUGUCCUGGCCCAAAAGGUAGCCAGUUGAAG_GGGUAGGAAGUGGG .......((((((((...........((((((((.((.........)).)))))))).(((((((....))..))))).......))))))))...... (-26.66 = -27.82 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:30 2006