| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,907,941 – 18,908,054 |
| Length | 113 |
| Max. P | 0.939649 |

| Location | 18,907,941 – 18,908,054 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.76 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -22.52 |
| Energy contribution | -22.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
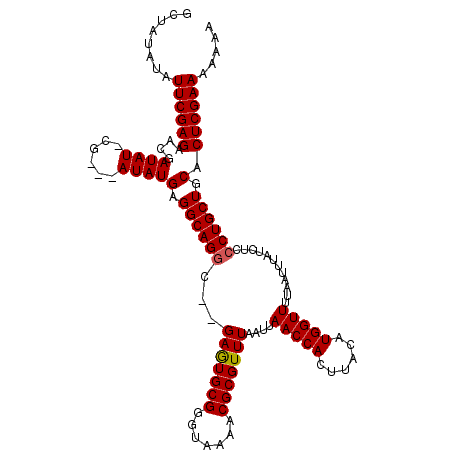
>2L_DroMel_CAF1 18907941 113 + 22407834 GCUAUAUAUUCGAGAACGAUAUGCG---AUAUGAGGCAGGCGCGAGUGCGGGUAAAACGCGUUUAAUUAACCACUUACAUGGUUUUAAUUUAUCUCCCUGCUGCACUCGAAAAAAA ........((((((..((.....))---...((.((((((.(.((..(((.......)))........(((((......)))))........)).))))))).))))))))..... ( -26.70) >DroSec_CAF1 33086 114 + 1 GCUAUAUAUUCGAGAACGAUAUACGAUGAUAUGAGGCAGGC--GAGUGCGGGUAAAACGCGUUUAAUUAACCACUUACAUGGUUUUAAUUUAUCUCCCUGCUGCACUCGAAAAAAA ........((((((..((.....))......((.((((((.--(((.(((.......)))........(((((......))))).........))))))))).))))))))..... ( -28.70) >DroSim_CAF1 32650 114 + 1 GCUAUAUAUUCGAGAACGAUAUGCGAUGAUAUGAGGCAGGC--GAAUGCGGGUAAAACGCGUUUAAUUAACCACUUACAUGGUUUUAAUUUAUCUCCCUGCUGCACUCGAAAAAAA ........((((((..((.....))......((.((((((.--(((((((.......)))))))....(((((......)))))............)))))).))))))))..... ( -28.70) >DroEre_CAF1 31264 104 + 1 GCUAUAUAUUCGAGAACGAU----------AUGAGGCAGUC--GAGUGCGGGUAAAACGCGUUUAAUUAACCACUUACAUGGUUUUAAUUUAUCUCCCUGCUGCACUCGAAAAAAA (((.(((((.((....))))----------))).)))..((--((((((((((.....))........(((((......)))))................))))))))))...... ( -26.30) >DroYak_CAF1 31950 104 + 1 GCUAUAUAUUCGAGAACGAU----------AUGAGGCAGGC--GAGUGCGGGUAAAACGCGUUUAAUUAACCACUUACAUGGUUUUAAUUUAUCUCCCUGCUGCACUCGAAAAAAU ........((((((......----------.((.((((((.--(((.(((.......)))........(((((......))))).........))))))))).))))))))..... ( -26.81) >consensus GCUAUAUAUUCGAGAACGAUAU_CG___AUAUGAGGCAGGC__GAGUGCGGGUAAAACGCGUUUAAUUAACCACUUACAUGGUUUUAAUUUAUCUCCCUGCUGCACUCGAAAAAAA ........((((((....((((......))))(.((((((...(((((((.......)))))))....(((((......)))))............)))))).).))))))..... (-22.52 = -22.76 + 0.24)


| Location | 18,907,941 – 18,908,054 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.76 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18907941 113 - 22407834 UUUUUUUCGAGUGCAGCAGGGAGAUAAAUUAAAACCAUGUAAGUGGUUAAUUAAACGCGUUUUACCCGCACUCGCGCCUGCCUCAUAU---CGCAUAUCGUUCUCGAAUAUAUAGC ........(((.((((..(.(((.........((((((....))))))........(((.......))).))).)..)))))))....---......(((....)))......... ( -25.50) >DroSec_CAF1 33086 114 - 1 UUUUUUUCGAGUGCAGCAGGGAGAUAAAUUAAAACCAUGUAAGUGGUUAAUUAAACGCGUUUUACCCGCACUC--GCCUGCCUCAUAUCAUCGUAUAUCGUUCUCGAAUAUAUAGC .....((((((.((.((((((((.........((((((....))))))........(((.......))).)))--.)))))...((((....))))...)).))))))........ ( -29.50) >DroSim_CAF1 32650 114 - 1 UUUUUUUCGAGUGCAGCAGGGAGAUAAAUUAAAACCAUGUAAGUGGUUAAUUAAACGCGUUUUACCCGCAUUC--GCCUGCCUCAUAUCAUCGCAUAUCGUUCUCGAAUAUAUAGC .....((((((.((.((((((((.........((((((....))))))........(((.......))).)))--.)))))...((((......)))).)).))))))........ ( -26.30) >DroEre_CAF1 31264 104 - 1 UUUUUUUCGAGUGCAGCAGGGAGAUAAAUUAAAACCAUGUAAGUGGUUAAUUAAACGCGUUUUACCCGCACUC--GACUGCCUCAU----------AUCGUUCUCGAAUAUAUAGC ........(((.((((..(((...........((((((....))))))........(((.......))).)))--..)))))))..----------.(((....)))......... ( -24.40) >DroYak_CAF1 31950 104 - 1 AUUUUUUCGAGUGCAGCAGGGAGAUAAAUUAAAACCAUGUAAGUGGUUAAUUAAACGCGUUUUACCCGCACUC--GCCUGCCUCAU----------AUCGUUCUCGAAUAUAUAGC .....((((((.((.((((((((.........((((((....))))))........(((.......))).)))--.))))).....----------...)).))))))........ ( -26.90) >consensus UUUUUUUCGAGUGCAGCAGGGAGAUAAAUUAAAACCAUGUAAGUGGUUAAUUAAACGCGUUUUACCCGCACUC__GCCUGCCUCAUAU___CG_AUAUCGUUCUCGAAUAUAUAGC .....((((((.((.((((((((.........((((((....))))))........(((.......))).)))...)))))...((((......)))).)).))))))........ (-23.56 = -23.80 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:28 2006