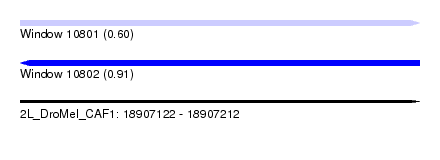
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,907,122 – 18,907,212 |
| Length | 90 |
| Max. P | 0.907519 |

| Location | 18,907,122 – 18,907,212 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -25.53 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
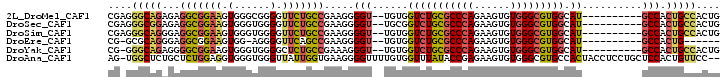
>2L_DroMel_CAF1 18907122 90 + 22407834 CGAGGGCAGAGAGGCGGAAGUGGGCGGGGUUCUGCCGAAGGGGU--UGUGGUCUGCGCCCAGAAGUGUGGGCGUGGCAU----------GCCACUGCCACUG ..(((((((...(((((((.(......).))))))).....(((--....(((.(((((((......))))))))))..----------))).))))).)). ( -39.80) >DroSec_CAF1 32264 90 + 1 CGAGGGCGGAGAGGCGGAAGUGGGUGGGGUUCUGCCGAAGGGGU--UGCGGUCUGCGCCCAGAAGUGUGGGCGUGGCAU----------GCCACUGCCACUG ..(((((((...(((((((.(......).))))))).....(((--....(((.(((((((......))))))))))..----------))).))))).)). ( -39.10) >DroSim_CAF1 31818 90 + 1 CGAGGGCAGGGAGGCGGAAGUGGGUGGGGUUCUGCCGAAGGGGU--UGUGGUCUGCGCCCAGAAGUGUGGGCGUGGCAU----------GCCACUGCCACUG ..(((((((...(((((((.(......).))))))).....(((--....(((.(((((((......))))))))))..----------))).))))).)). ( -40.00) >DroEre_CAF1 30473 82 + 1 CG-GCGCAGGGAGGCGGAAGUGG-AGGGGUUCAGCCGAAGGGGU--UGUGGUCUGCGCCCAGAAGUGUGGGCGUGGCAU----------GCCACUG------ ((-((........((....))((-(....))).)))).......--.((((((..((((((......))))))..)...----------)))))..------ ( -30.60) >DroYak_CAF1 31163 89 + 1 CG-GGGCAGAGGGGCGGAAGUGGGUGGGGCUCUGCCGAAAGGGU--UGUGGUCUGCGCCCAGAAGUGUGGGCGUGGCAU----------GCCACUGCCACUG ((-((((((((..((....))........))))))).....(((--.((((((..((((((......))))))..)...----------))))).))).))) ( -40.20) >DroAna_CAF1 17770 99 + 1 AG-UGGCUCUGCUCUGGAGGUGGGUGGGUUAUUGGUGAAGGGGUUUUGUGGUUUAUACCGAGAAGUGUGGGCGUGCCACUACCUCCUGCUCCACUGUUCC-- ((-(((....((...((((((((.(((.......(..(((...)))..).((((((((......))))))))...))))))))))).)).))))).....-- ( -31.30) >consensus CG_GGGCAGAGAGGCGGAAGUGGGUGGGGUUCUGCCGAAGGGGU__UGUGGUCUGCGCCCAGAAGUGUGGGCGUGGCAU__________GCCACUGCCACUG ....(((((...(((((((.(......).))))))).....(((......(((((((((((......))))))))).))..........))).))))).... (-25.53 = -26.12 + 0.59)
| Location | 18,907,122 – 18,907,212 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -17.30 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
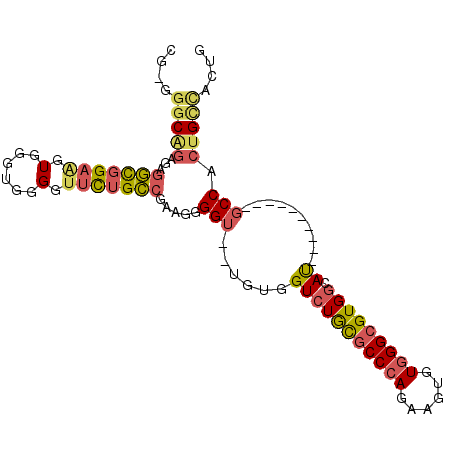
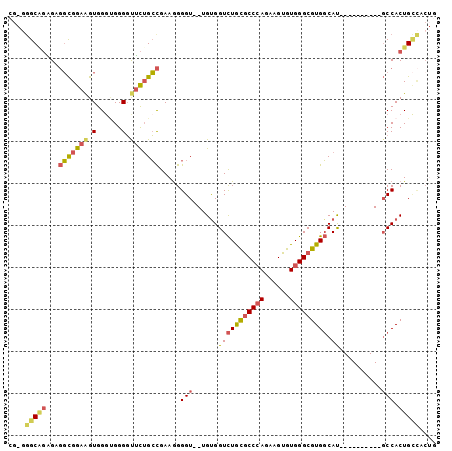
>2L_DroMel_CAF1 18907122 90 - 22407834 CAGUGGCAGUGGC----------AUGCCACGCCCACACUUCUGGGCGCAGACCACA--ACCCCUUCGGCAGAACCCCGCCCACUUCCGCCUCUCUGCCCUCG ..(((((.((((.----------...))))(((((......)))))...).)))).--........((((((.....((........))...)))))).... ( -30.70) >DroSec_CAF1 32264 90 - 1 CAGUGGCAGUGGC----------AUGCCACGCCCACACUUCUGGGCGCAGACCGCA--ACCCCUUCGGCAGAACCCCACCCACUUCCGCCUCUCCGCCCUCG ..((((.((.(((----------.((((..(((((......)))))((.....)).--........))))(((..........))).))).))))))..... ( -28.30) >DroSim_CAF1 31818 90 - 1 CAGUGGCAGUGGC----------AUGCCACGCCCACACUUCUGGGCGCAGACCACA--ACCCCUUCGGCAGAACCCCACCCACUUCCGCCUCCCUGCCCUCG .((.((((((((.----------.(((...(((((......))))))))..)))).--........(((.(((..........))).)))....)))))).. ( -29.00) >DroEre_CAF1 30473 82 - 1 ------CAGUGGC----------AUGCCACGCCCACACUUCUGGGCGCAGACCACA--ACCCCUUCGGCUGAACCCCU-CCACUUCCGCCUCCCUGCGC-CG ------..((((.----------.(((...(((((......))))))))..)))).--.......((((.((.....)-).......((......))))-)) ( -23.10) >DroYak_CAF1 31163 89 - 1 CAGUGGCAGUGGC----------AUGCCACGCCCACACUUCUGGGCGCAGACCACA--ACCCUUUCGGCAGAGCCCCACCCACUUCCGCCCCUCUGCCC-CG ..(((((.((((.----------...))))(((((......)))))...).)))).--........(((((((..................))))))).-.. ( -31.67) >DroAna_CAF1 17770 99 - 1 --GGAACAGUGGAGCAGGAGGUAGUGGCACGCCCACACUUCUCGGUAUAAACCACAAAACCCCUUCACCAAUAACCCACCCACCUCCAGAGCAGAGCCA-CU --.....(((((.((.((((((.((((.....)))).......(((....)))............................))))))...))....)))-)) ( -24.20) >consensus CAGUGGCAGUGGC__________AUGCCACGCCCACACUUCUGGGCGCAGACCACA__ACCCCUUCGGCAGAACCCCACCCACUUCCGCCUCUCUGCCC_CG ..((((.(((((............((((.((((((......))))))..((.............)))))).........))))).))))............. (-17.30 = -19.22 + 1.92)

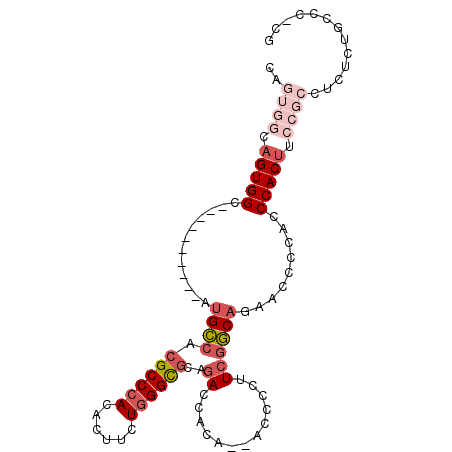
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:26 2006