
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,904,533 – 18,904,628 |
| Length | 95 |
| Max. P | 0.970143 |

| Location | 18,904,533 – 18,904,628 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.78 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -6.66 |
| Energy contribution | -6.33 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.23 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
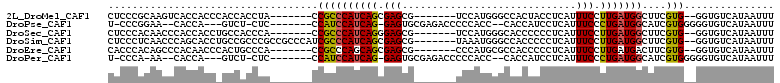
>2L_DroMel_CAF1 18904533 95 - 22407834 CUCCCGCAAGUCACCACCCACCACCUA-------CCGCCCAUCAGCGAGCG-------UCCAUGGGCCACUACCUCAUUUCCUUGAUGGCUUCGUG--GGUGUCAUAAUUU ....(....)....(((((((......-------..((((((..((....)-------)..)))))).....(((((......))).))....)))--))))......... ( -26.70) >DroPse_CAF1 25976 94 - 1 U-CCCGGAA--CACCA---GUCU-CUC-------CCAUCCAUCAG-GAGUGCGAGACCCCCACC--CACCAUCCUCAUUUCCCUGAUGGCAUCGUGGGGGUGUCAUAAUUU .-...((..--..)).---((..-(((-------(.........)-))).))((.((((((((.--..(((((...........)))))....)))))))).))....... ( -30.40) >DroSec_CAF1 29728 95 - 1 CUCCCACAACCCACCACCUGCCACCCA-------CCGCCCAUCAGGGAGCG-------UCCAUGGGCACCCCCCUCAUUUCCUUGAUGGCUUCGUG--GGUGUCAUAAUUU ..................((.((((((-------(...(((((((((((..-------.....(((.....)))....)))))))))))....)))--)))).))...... ( -32.00) >DroSim_CAF1 28914 102 - 1 CUCCCUCAACCCAGCACCUGCCGCCCGCCGCCCAUCGCCCAUCAGCGAGCG-------UAAAUGGGCCACCCCCUCAUUUCCUUGAUGGCUUCGUG--GGUGUCAUAAUUU .............(((((..(.(((((.(((...((((......)))))))-------....)))))....((.(((......))).))....)..--)))))........ ( -28.40) >DroEre_CAF1 27937 95 - 1 CACCCACAGCCCACAACCCACUGCCCA-------CCGCCCAGCAGCGAGCG-------CCCAUGCGCCACCCCCUCAUUUCCUUGAUGACUUCGUG--GGUGUCAUAAUUU (((((((...........(.((((...-------.......)))).).(((-------(....)))).......(((((.....)))))....)))--))))......... ( -24.30) >DroPer_CAF1 28034 93 - 1 U-CCCA-AA--CACCA---GUCU-CUC-------CCAUCCAUCAG-GAGUGCGAGACCCCCACC--CACCAUCCUCAUUUCCCUGAUGGCAUCGUGGGGGUGUCAUAAUUU .-....-..--.....---((..-(((-------(.........)-))).))((.((((((((.--..(((((...........)))))....)))))))).))....... ( -29.60) >consensus CUCCCACAACCCACCACC_GCCACCCA_______CCGCCCAUCAGCGAGCG_______CCCAUGGGCAACCCCCUCAUUUCCUUGAUGGCUUCGUG__GGUGUCAUAAUUU ...................................((((((((((.(((.............................))).)))))))....)))............... ( -6.66 = -6.33 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:24 2006