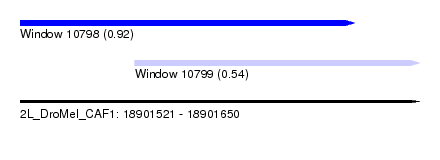
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,901,521 – 18,901,650 |
| Length | 129 |
| Max. P | 0.924246 |

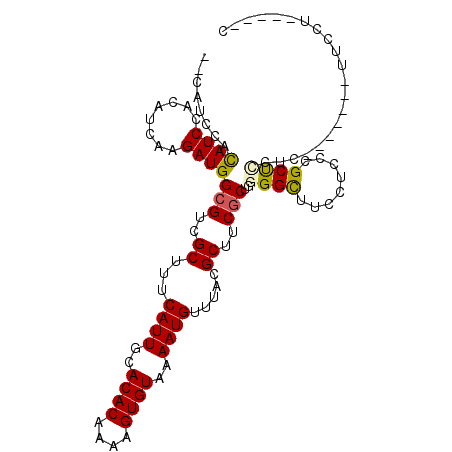
| Location | 18,901,521 – 18,901,629 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18901521 108 + 22407834 -UCAUUCACAUCCACAUCAAGAUGGCGUCGCUUUCAUUGCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCCUCCCGCUUCUUCCAUCCUUC -...................(((((((..((...((((..((((....))))..)))).....))..))).(((((........)))))............)))).... ( -23.10) >DroPse_CAF1 22698 92 + 1 ----UCCCCAUCAACAUAAAGAUGGCGUCGCUUUCAUUGCACACAAAAGUGUAAAAUGUUUACGCUUCGCUUAGGUUUCCUUUUGCCGCUU-------UUCCU------ ----....((((........))))(((..((...((((..((((....))))..)))).....))..)))...(((........)))....-------.....------ ( -18.20) >DroSim_CAF1 25900 105 + 1 -CCAUUCACAUCCACAUCAAGAUGGCGUCGCCUUCAUUGCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCCUCC---AUCUUCCUUUCUUC -.................(((((((((..((...((((..((((....))))..)))).....))..))).(((((........)))))..)---)))))......... ( -25.00) >DroYak_CAF1 25516 106 + 1 CACAUCCACAUCCACAUCAAGAUGGCGUCGCUUUCAUUGCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCGUCC---UGCUUCCUUCCUUC ........((((........))))(((..((...((((..((((....))))..)))).....))..))).((((.....))))((......---.))........... ( -20.40) >DroAna_CAF1 13631 88 + 1 -ACAUCCAUAUCCCCAUCAAGAUGGCGUCGCUUUCAUUGCACACAAAAGUGUAAAAUGUUUACGCUUCACUUGGGCUUCCUUUAGCCU--------------------C -................((((..(((((......((((..((((....))))..))))...)))))...))))((((......)))).--------------------. ( -20.60) >DroPer_CAF1 24606 92 + 1 ----UCCCCAUCAACAUAAAGAUGGCGUCGCUUUCAUUGCACACAAAAGUGUAAAAUGUUUACGCUUCGCUUAGGUUUCCUUUUGCUGCUU-------UUCCU------ ----...(((((........))))).............(((.....((((((((.....)))))))).((..(((...)))...)))))..-------.....------ ( -16.60) >consensus __CAUCCACAUCCACAUCAAGAUGGCGUCGCUUUCAUUGCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCCUC_______UUCCU_____C ........((((........))))(((..((...((((..((((....))))..)))).....))..)))..((((........))))..................... (-16.60 = -16.43 + -0.17)

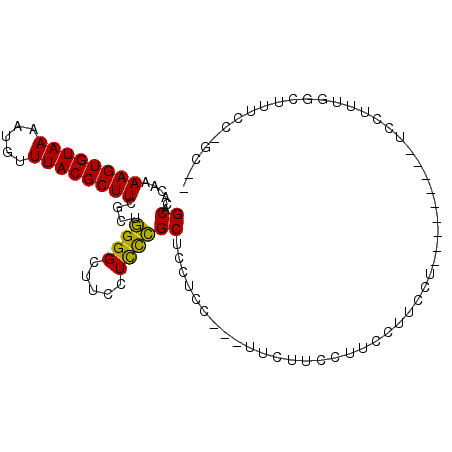
| Location | 18,901,558 – 18,901,650 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.82 |
| Mean single sequence MFE | -16.86 |
| Consensus MFE | -11.38 |
| Energy contribution | -10.74 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18901558 92 + 22407834 GCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCCUCCCGCUUCUUCCAUCCUUCCU---------GCCUUUAGCUUUCCCGCUG (((.....((((((((.....)))))))).((.(((((........)))))....))...............)---------))...((((......)))) ( -18.30) >DroSim_CAF1 25937 89 + 1 GCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCCUCC---AUCUUCCUUUCUUCCU---------UCCUUUGGCUUUCCCGCUG ((......((((((((.....)))))))).(..(((((........)))))..)---................---------...............)).. ( -15.70) >DroEre_CAF1 24958 90 + 1 GCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCGUCC---UUCUUCCUUCCCUCC-CGCUGCCCAUCCUCUGGCUUU------- ((......((((((((.....)))))))).((.((((....)))).))......---...............-.)).(((........)))...------- ( -17.50) >DroYak_CAF1 25554 91 + 1 GCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCGUCC---UGCUUCCUUCCUUCCUCACUGCCCAUCCUUUGGCCUU------- (((.((..((((((((.....)))))))).((.((((....)))).))..))..---))).................(((........)))...------- ( -17.40) >DroAna_CAF1 13668 72 + 1 GCACACAAAAGUGUAAAAUGUUUACGCUUCACUUGGGCUUCCUUUAGCCU--------------------CCC---------CCCUUUGUCCGGCCGGCCA ((..((((((((((((.....)))))))).....(((((......)))))--------------------...---------....))))...))...... ( -15.40) >consensus GCACACAAAAGUGUAAAAUGUUUACGCUUCGCUGGGGCUUCCUCCCGCUCCUCC___UUCUUCCUUCCUUCCU_________UCCUUUGGCUUUCC_GC__ ((......((((((((.....))))))))....((((.....))))))..................................................... (-11.38 = -10.74 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:23 2006