| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,975,029 – 1,975,145 |
| Length | 116 |
| Max. P | 0.722761 |

| Location | 1,975,029 – 1,975,145 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
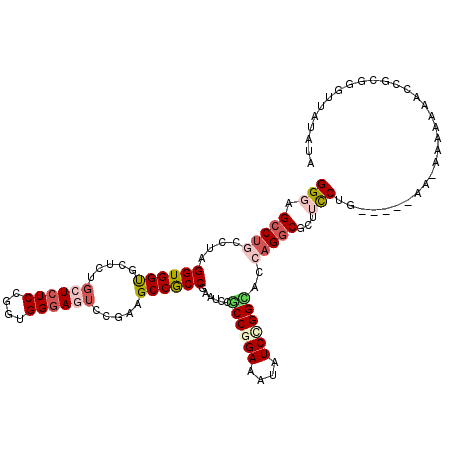
| Mean single sequence MFE | -40.89 |
| Consensus MFE | -28.98 |
| Energy contribution | -30.28 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
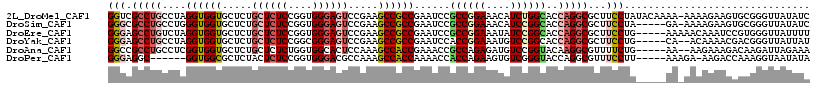
>2L_DroMel_CAF1 1975029 116 - 22407834 GGUCGCCUGCCUAGGUGGUGCUCUGCUCUCCGGUGGGAGUCCGAAGCCGCCGAAUCCGCCGGAAACAUCUGGCACCAGGCGCUUCCUAUACAAAA-AAAAGAAGUGCGGGUUAUAUC (((.....)))..((((((..((.((((((....))))))..)).)))))).(((((((((((....)))))).....(((((((..........-....))))))))))))..... ( -42.34) >DroSim_CAF1 774 111 - 1 GGGCGCCUGCCUGGGUGGUGCUCUGCUCUCCGGUGGGAGUCCGAAGCCGCCGAAUCCGCCGGAAACAUCCGGCACCAGGCGCUUCCUA-----GA-AAAAGAAGUGCGGGUUAUAUC ((((....)))).((((((..((.((((((....))))))..)).)))))).(((((((((((....)))))).....(((((((...-----..-....))))))))))))..... ( -47.90) >DroEre_CAF1 873 112 - 1 GGGAGCCUGUCUAGGUGGUGCUCUGCUCUCCGGUGGGAGUCCGAAGCCGCCGAAUCCGCCGGAAAUAUCCGGCACCAGGCGCUUCCUG-----AAAAACAAAUCCGUGGGUUAUUUU ((((((..((((.((((((..((.((((((....))))))..)).))))))......((((((....))))))...))))))))))..-----.......((((....))))..... ( -43.10) >DroYak_CAF1 768 110 - 1 GGGAGCCUGCCUAGGUGGUGCUCUGCUCUCCGGCGGGAGUCCGAAGCCGCCGAAUCCACCGGAAAUGUCCGGCACCAGGCGCUUCCUG-----CA--ACAAAACGACGGGUUAUUAU ((((((..((((.((((((..((.((((((....))))))..)).)))))).......(((((....)))))....))))))))))..-----..--.................... ( -39.80) >DroAna_CAF1 1035 110 - 1 GGCCGCCUGCCUCGGUGGUGCUCUGCUCUCUGGUGGCACUCCAAAGCCACCGAAACCGCCAGAGAUGUCCGGUACAAGGCGUUUUCUG-----AA--AAGAAAGACAAGAUUAGAAA .((((((......))).(((((..((((((((((((...................)))))))))).))..)))))..)))(((((((.-----..--.)).)))))........... ( -36.31) >DroPer_CAF1 16427 105 - 1 GGGAGGC------GGUGGCGCUCUACUCUCCGGUGGGACGCCAAAGCCACCAAAACCACCAGAAGUGUCGGGUACCAGGCGUUUCCUU-----AAAGA-AAGACCAAAGGUAAUAUA .((.(((------..(((((..(((((....)))))..)))))..))).))...((((((.((....)).)))....((..((((...-----...))-))..))...)))...... ( -35.90) >consensus GGGAGCCUGCCUAGGUGGUGCUCUGCUCUCCGGUGGGAGUCCGAAGCCGCCGAAUCCGCCGGAAAUAUCCGGCACCAGGCGCUUCCUG_____AA_AAAAAAACCGCGGGUUAUAUA (((.(((((....((((((.....((((((....)))))).....))))))......((((((....))))))..)))))...)))............................... (-28.98 = -30.28 + 1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:37 2006