| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,877,267 – 18,877,365 |
| Length | 98 |
| Max. P | 0.899229 |

| Location | 18,877,267 – 18,877,365 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -22.59 |
| Energy contribution | -24.32 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18877267 98 + 22407834 CACCCCGAAAUGUAUC-UCGCAGAUACAUUCUGCCUGCGAGAGCAGUGCGACUAGCUCGUCGCUCCGCAAGGGUAGGGAACUGGUGUGGC--ACUGGUGCA (((.(((.((((((((-.....))))))))((((((((....)))).(((((......))))).((....)))))).....))).)))((--(....))). ( -37.60) >DroGri_CAF1 1232 86 + 1 C--CUUCCACAU-UGUUGAGUUGCUUC-UU----GUGUUAGAGCAGCACAGCUAGCGGG-GGUU-UGCUAG----AUGUGCGUGUGUGAGUGUGUG-UGCG .--((..(((((-........((((.(-(.----.....))))))(((((.(((((((.-...)-))))))----.))))))))))..))......-.... ( -27.00) >DroSec_CAF1 744 97 + 1 CACCCCGAAAUGUAUC-UCGCAGAUAC-UUCUGCCUGCGAGAGCAGUGCGACUAGCUCGUCGCUCCGCAAGGGUAGGGUACUGGUGUGGC--ACUGGUGCA .((((.........((-((((((....-......))))))))((.(.(((((......))))).).))..))))...((((..(((...)--))..)))). ( -40.00) >DroSim_CAF1 744 97 + 1 CACCCCGAAAUGUAUC-UCGCAGAUAC-UUCUGCCUGCGAGAGCAGUGCGACUAGCUCUUCGCUCCGCAAGGGUAGGGUACUGGUGUGGC--ACUGGUGCA ((((.......((.((-((((((....-......))))))))))(((((.(((((..((..((.((....))))..))..)))))...))--))))))).. ( -38.20) >DroEre_CAF1 680 97 + 1 CACCCCGAAAUGUAUC-UCGCAGAUAC-UUCUGCCUGCGAGAGCAGUGCGACUAGCUCGUCGCUCCGCAAGGGUAAGGUACUGGUAUGGC--ACUGGUGCA .((((.........((-((((((....-......))))))))((.(.(((((......))))).).))..))))...((((..((.....--))..)))). ( -38.30) >DroYak_CAF1 748 97 + 1 CACCCCGAAAAGUAUC-UCGCAGAUAC-UUCUGCCUGCGAGAGCAGUGCGACUAGCUCGUCGCUCCGCAAGGGUAAGGUACUGGUGUGGC--ACUGGUGCA .((((.........((-((((((....-......))))))))((.(.(((((......))))).).))..))))...((((..(((...)--))..)))). ( -40.00) >consensus CACCCCGAAAUGUAUC_UCGCAGAUAC_UUCUGCCUGCGAGAGCAGUGCGACUAGCUCGUCGCUCCGCAAGGGUAAGGUACUGGUGUGGC__ACUGGUGCA .((((..............(((((.....)))))((((....)))).(((((......))))).......))))...((((..((.......))..)))). (-22.59 = -24.32 + 1.72)



| Location | 18,877,267 – 18,877,365 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -20.68 |
| Energy contribution | -22.43 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18877267 98 - 22407834 UGCACCAGU--GCCACACCAGUUCCCUACCCUUGCGGAGCGACGAGCUAGUCGCACUGCUCUCGCAGGCAGAAUGUAUCUGCGA-GAUACAUUUCGGGGUG .(((....)--)).............((((((.((((.(((((......))))).))))((((((((((.....))..))))))-))........)))))) ( -36.40) >DroGri_CAF1 1232 86 - 1 CGCA-CACACACUCACACACGCACAU----CUAGCA-AACC-CCCGCUAGCUGUGCUGCUCUAACAC----AA-GAAGCAACUCAACA-AUGUGGAAG--G ....-......((..((((.(((((.----(((((.-....-...))))).)))))((((((.....----.)-).))))........-.))))..))--. ( -25.10) >DroSec_CAF1 744 97 - 1 UGCACCAGU--GCCACACCAGUACCCUACCCUUGCGGAGCGACGAGCUAGUCGCACUGCUCUCGCAGGCAGAA-GUAUCUGCGA-GAUACAUUUCGGGGUG .(((....)--))........((((((......((((.(((((......))))).))))(((((((((.....-...)))))))-))........)))))) ( -37.70) >DroSim_CAF1 744 97 - 1 UGCACCAGU--GCCACACCAGUACCCUACCCUUGCGGAGCGAAGAGCUAGUCGCACUGCUCUCGCAGGCAGAA-GUAUCUGCGA-GAUACAUUUCGGGGUG .(((....)--))........((((((......((((.((((........)))).))))(((((((((.....-...)))))))-))........)))))) ( -33.40) >DroEre_CAF1 680 97 - 1 UGCACCAGU--GCCAUACCAGUACCUUACCCUUGCGGAGCGACGAGCUAGUCGCACUGCUCUCGCAGGCAGAA-GUAUCUGCGA-GAUACAUUUCGGGGUG .(((....)--)).............((((((.((((.(((((......))))).))))(((((((((.....-...)))))))-))........)))))) ( -37.10) >DroYak_CAF1 748 97 - 1 UGCACCAGU--GCCACACCAGUACCUUACCCUUGCGGAGCGACGAGCUAGUCGCACUGCUCUCGCAGGCAGAA-GUAUCUGCGA-GAUACUUUUCGGGGUG .(((....)--)).............((((((.((((.(((((......))))).))))(((((((((.....-...)))))))-))........)))))) ( -37.10) >consensus UGCACCAGU__GCCACACCAGUACCCUACCCUUGCGGAGCGACGAGCUAGUCGCACUGCUCUCGCAGGCAGAA_GUAUCUGCGA_GAUACAUUUCGGGGUG ..........................((((((.((((.(((((......))))).))))(((((((((.........))))))).))........)))))) (-20.68 = -22.43 + 1.75)


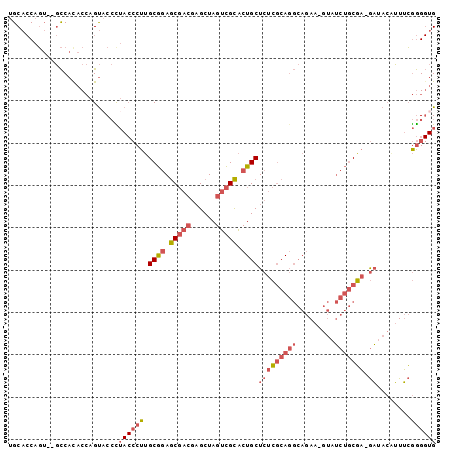
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:15 2006