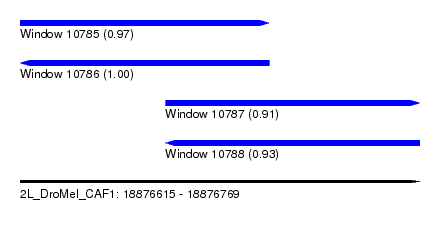
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,876,615 – 18,876,769 |
| Length | 154 |
| Max. P | 0.998278 |

| Location | 18,876,615 – 18,876,711 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -31.31 |
| Energy contribution | -31.00 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18876615 96 + 22407834 CCAGGUGAUGGUUCAUCACCAGCGAUGGCUGUAAUCCAGUUU------------------------GAUUGUGGUUGUGAUUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCU ...((((........((((((((.(..((..(((((......------------------------)))))..))..).((((.....)))).))))))))(((....)))))))..... ( -35.40) >DroGri_CAF1 249 96 + 1 CCAGGUGAUGAUUCAUCACCAGCGAUGGCUGUAAUCCAGUUU------------------------GAUUGUGAUUGUGGUUGUGCAAUGGUAGCUGGUGAGCCAAAUGGCCACCAAUCU ((((((((((...))))))).((.(..((..(((((((((..------------------------.)))).)))))..))..)))..)))....(((((.(((....)))))))).... ( -38.40) >DroEre_CAF1 30 96 + 1 CCAGGUGAUGGUUCAUCACCAGCGAUGGCUGUAAUCCAGUUU------------------------GAUUGUGGUUGUGAUUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCU ...((((........((((((((.(..((..(((((......------------------------)))))..))..).((((.....)))).))))))))(((....)))))))..... ( -35.40) >DroYak_CAF1 108 96 + 1 CCAGGUGAUGGUUCAUCACCAGCGAUGGCUGUAAUCCAGUUU------------------------GAUUGUGGUUGUGAUUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCU ...((((........((((((((.(..((..(((((......------------------------)))))..))..).((((.....)))).))))))))(((....)))))))..... ( -35.40) >DroMoj_CAF1 30 120 + 1 CCAGGUGAUGAUUCAUCACCAGCGAUGGCUGUAAUCCAGUUUGGCUGUGACUGUGAUUGUGAUUGUGAUUGUGAUUAUGAUUGUGCAAUGGUAGCUGGUGAGCCAAAUGGCCACCAAUCU ...((((........((((((((....((((((((((((((.......))))).))))(..((..((((....))))..))..)...))))).))))))))(((....)))))))..... ( -39.90) >DroPer_CAF1 30 96 + 1 CCAGGUGAUGGUUCAUCACCAGCGACGGCUGUAAUCCAGUUU------------------------GAUUGUGGUUGUGAUUGUGCAAUGGUAGCUGGUGAGCCAAAUGGCCACCAAUCU ...((((........((((((((.(((((..(((((......------------------------)))))..))))).....(((....)))))))))))(((....)))))))..... ( -36.60) >consensus CCAGGUGAUGGUUCAUCACCAGCGAUGGCUGUAAUCCAGUUU________________________GAUUGUGGUUGUGAUUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCU ...((((........((((((((...(((((.....)))))................................(((((......)))))....))))))))(((....)))))))..... (-31.31 = -31.00 + -0.31)

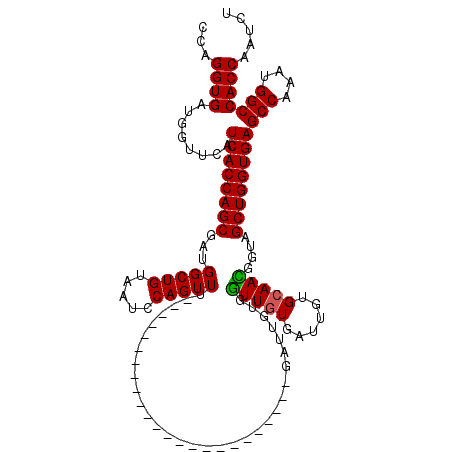

| Location | 18,876,615 – 18,876,711 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.35 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18876615 96 - 22407834 AGAUUGGUGGCCAUUUGGCUCACCAGCUACCGUUGCACAAUCACAACCACAAUC------------------------AAACUGGAUUACAGCCAUCGCUGGUGAUGAACCAUCACCUGG .....(((((((....)))((((((((....((((........)))).......------------------------...(((.....))).....))))))))........))))... ( -28.10) >DroGri_CAF1 249 96 - 1 AGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAACCACAAUCACAAUC------------------------AAACUGGAUUACAGCCAUCGCUGGUGAUGAAUCAUCACCUGG .(((((((((((....))).)))).((.......))........))))......------------------------...(..(....((((....))))((((((...)))))))..) ( -26.50) >DroEre_CAF1 30 96 - 1 AGAUUGGUGGCCAUUUGGCUCACCAGCUACCGUUGCACAAUCACAACCACAAUC------------------------AAACUGGAUUACAGCCAUCGCUGGUGAUGAACCAUCACCUGG .....(((((((....)))((((((((....((((........)))).......------------------------...(((.....))).....))))))))........))))... ( -28.10) >DroYak_CAF1 108 96 - 1 AGAUUGGUGGCCAUUUGGCUCACCAGCUACCGUUGCACAAUCACAACCACAAUC------------------------AAACUGGAUUACAGCCAUCGCUGGUGAUGAACCAUCACCUGG .....(((((((....)))((((((((....((((........)))).......------------------------...(((.....))).....))))))))........))))... ( -28.10) >DroMoj_CAF1 30 120 - 1 AGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAAUCAUAAUCACAAUCACAAUCACAAUCACAGUCACAGCCAAACUGGAUUACAGCCAUCGCUGGUGAUGAAUCAUCACCUGG .(((((((((((....))).)))).((.......))..)))).........................(((...((((....(((.....))).....))))((((((...))))))))). ( -30.10) >DroPer_CAF1 30 96 - 1 AGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAAUCACAACCACAAUC------------------------AAACUGGAUUACAGCCGUCGCUGGUGAUGAACCAUCACCUGG .(((((((((((....))).)))).((.......))..))))....(((.....------------------------...(((.....)))........(((((((...)))))))))) ( -27.30) >consensus AGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAAUCACAACCACAAUC________________________AAACUGGAUUACAGCCAUCGCUGGUGAUGAACCAUCACCUGG .....(((((((....)))((((((((....((((........))))..................................(((.....))).....))))))))........))))... (-26.66 = -26.35 + -0.31)


| Location | 18,876,671 – 18,876,769 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -23.50 |
| Energy contribution | -23.37 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18876671 98 + 22407834 UUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCUCUGCCAUUACCAUAUAUUUGGCUGACAGGA--------CACUCGGCGA-A-GCACACUCGAG-AUA-----------UUU .(((((...(((((.(((((.(((....))))))))....)))))............((.(((((.....--------...))))).)-)-)))))......-...-----------... ( -32.60) >DroGri_CAF1 305 108 + 1 UUGUGCAAUGGUAGCUGGUGAGCCAAAUGGCCACCAAUCUCUGCCAUUACCAUAUAUUUGGCUUAUAGACCCCGGCUUAACCACCCAAAA-ACACACUUGUGUGUC-----------UUA .((((.((((((((.(((((.(((....))))))))....))))))))..))))..(((((.....((.(....))).......))))).-(((((....))))).-----------... ( -30.40) >DroEre_CAF1 86 98 + 1 UUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCUCUGCCAUUACCAUAUAUUUGGCUGACAGGA--------CACUCGGCGA-A-ACACACUCGAG-AUU-----------UUU .((((....(((((.(((((.(((....))))))))....)))))...........(((.(((((.....--------...))))).)-)-)))))......-...-----------... ( -28.90) >DroYak_CAF1 164 98 + 1 UUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCUCUGCCAUUACCAUAUAUUUGGCUGACAGGA--------CACUCGGCGA-A-GCACACUCGAG-AUU-----------UUU .(((((...(((((.(((((.(((....))))))))....)))))............((.(((((.....--------...))))).)-)-)))))......-...-----------... ( -32.60) >DroMoj_CAF1 110 102 + 1 UUGUGCAAUGGUAGCUGGUGAGCCAAAUGGCCACCAAUCUCUGCCAUUACCAUAUAUUUGGCUUAUAGAC--CGGC--AACCAACCAA-A-AUACACUUCAG-AUC-----------UUU ...(((((((((((.(((((.(((....))))))))....)))))))).(((......))).........--..))--).........-.-...........-...-----------... ( -26.70) >DroPer_CAF1 86 109 + 1 UUGUGCAAUGGUAGCUGGUGAGCCAAAUGGCCACCAAUCUCUGCCAUUACCAUAUAUUUGGCUUACAAAA--------CC-CCGGCAA-ACACACACUCGAG-ACGGGGUGGGUGGGUGU .((((.((((((((.(((((.(((....))))))))....))))))))..))))......((((((...(--------((-(((....-.............-.))))))..)))))).. ( -40.77) >consensus UUGUGCAACGGUAGCUGGUGAGCCAAAUGGCCACCAAUCUCUGCCAUUACCAUAUAUUUGGCUGACAGAA________CACCCGGCAA_A_ACACACUCGAG_AUC___________UUU .((((....(((((.(((((.(((....))))))))....)))))....(((......)))...............................))))........................ (-23.50 = -23.37 + -0.14)
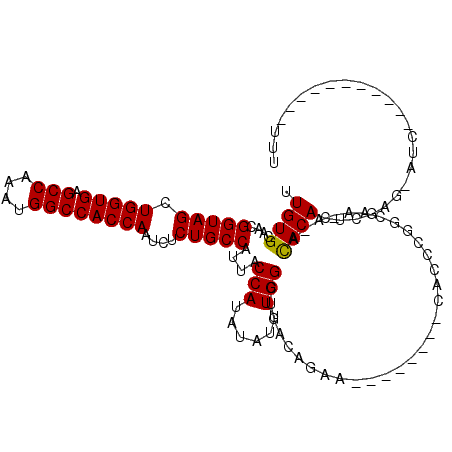
| Location | 18,876,671 – 18,876,769 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -26.85 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18876671 98 - 22407834 AAA-----------UAU-CUCGAGUGUGC-U-UCGCCGAGUG--------UCCUGUCAGCCAAAUAUAUGGUAAUGGCAGAGAUUGGUGGCCAUUUGGCUCACCAGCUACCGUUGCACAA ...-----------...-......(((((-.-.((...((((--------((((((((((((......))))..)))))).)))((((((((....))).))))))))..))..))))). ( -35.70) >DroGri_CAF1 305 108 - 1 UAA-----------GACACACAAGUGUGU-UUUUGGGUGGUUAAGCCGGGGUCUAUAAGCCAAAUAUAUGGUAAUGGCAGAGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAA .((-----------((((((....)))))-)))..(((...((..(....)..))...))).......(((((((((...((.(((((((((....))).)))))))).))))))).)). ( -37.10) >DroEre_CAF1 86 98 - 1 AAA-----------AAU-CUCGAGUGUGU-U-UCGCCGAGUG--------UCCUGUCAGCCAAAUAUAUGGUAAUGGCAGAGAUUGGUGGCCAUUUGGCUCACCAGCUACCGUUGCACAA ...-----------...-......(((((-.-.((...((((--------((((((((((((......))))..)))))).)))((((((((....))).))))))))..))..))))). ( -35.10) >DroYak_CAF1 164 98 - 1 AAA-----------AAU-CUCGAGUGUGC-U-UCGCCGAGUG--------UCCUGUCAGCCAAAUAUAUGGUAAUGGCAGAGAUUGGUGGCCAUUUGGCUCACCAGCUACCGUUGCACAA ...-----------...-......(((((-.-.((...((((--------((((((((((((......))))..)))))).)))((((((((....))).))))))))..))..))))). ( -35.70) >DroMoj_CAF1 110 102 - 1 AAA-----------GAU-CUGAAGUGUAU-U-UUGGUUGGUU--GCCG--GUCUAUAAGCCAAAUAUAUGGUAAUGGCAGAGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAA ...-----------...-.....(((((.-.-.((((((((.--.(((--((((....((((............))))..)))))))..))))...((((....)))))))).))))).. ( -35.30) >DroPer_CAF1 86 109 - 1 ACACCCACCCACCCCGU-CUCGAGUGUGUGU-UUGCCGG-GG--------UUUUGUAAGCCAAAUAUAUGGUAAUGGCAGAGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAA ..........((((((.-..((((......)-))).)))-))--------).................(((((((((...((.(((((((((....))).)))))))).))))))).)). ( -34.30) >consensus AAA___________AAU_CUCGAGUGUGU_U_UCGCCGAGUG________UCCUGUAAGCCAAAUAUAUGGUAAUGGCAGAGAUUGGUGGCCAUUUGGCUCACCAGCUACCAUUGCACAA .......................((((...............................((((......))))(((((...((.(((((((((....))).)))))))).))))))))).. (-26.85 = -26.60 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:13 2006