
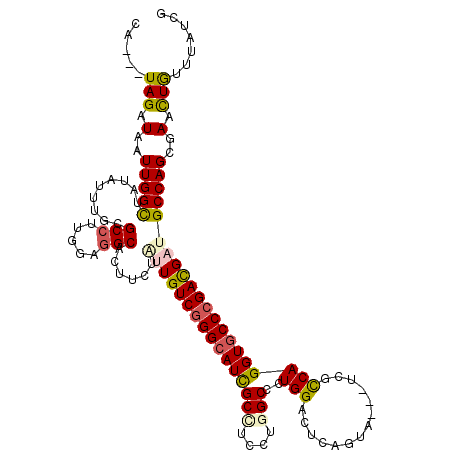
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,849,915 – 18,850,029 |
| Length | 114 |
| Max. P | 0.588422 |

| Location | 18,849,915 – 18,850,029 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18849915 114 + 22407834 AA---UAGAUAAUUGGCUAUCUGUGCGCCGUGGAAGCAUUUCUACUGUCGGGCAUCGCAUCCUGGCCGUGGACUCAGUA---CCGCCAGGUGCCCGACGAUGCCAGCGAAUUGUUCAUCG ..---..(((((((.(((....(.(((.(((((((....)))))).((((((((......((((((.(((.......))---).))))))))))))))).))))))).)))))))..... ( -41.10) >DroVir_CAF1 951 114 + 1 CU---UAGCUAAUUGGCUUUAUUUGCGCCUUGGAGGCGCUGCUGCUGUCGGGCAUUGCCUCCUGGCCCUGGUCUCAGUA---UCGCCAGGUGCCCGACGAUGCCAGCGAGCUGUUCAUUG ..---(((((..(((((.......(((((.....)))))......(((((((((..(((....)))((((((.......---..)))))))))))))))..)))))..)))))....... ( -51.60) >DroGri_CAF1 1234 114 + 1 CU---UAGAUAAUUGGCUAUAUUUGCGCCUUGGAGGCACUGCUGUUGUCCGGCAUUGCCUCGUGGCCCUGGUCCCAAUA---UCGUCAGGUGCCCGACGAUGCCAGCGAACUGUUUAUCG ..---..(((((..((.....((((((((...((((((.(((((.....))))).))))))..)))..((((......(---(((((.(....).)))))))))))))))))..))))). ( -40.10) >DroWil_CAF1 836 117 + 1 UA---UAGAUAAUUGGUUAUCUUUGCGCCUUGGAGGCAUUUCUAUUGUCGGGCAUUGCUUCGUGGCCGUGGAUUCAGUAUUAUCGUCAGGUUCCCGAUGAUGCCAGUGAACUAUUUAUUG ..---..(((((.(((((((((.((.(((...((((((..(((......)))...))))))..))))).))))...(((((((((.........))))))))).....))))).))))). ( -30.30) >DroMoj_CAF1 629 114 + 1 CU---UAGCUAAUUGGCUACAUUUGCGCCUUGGAGGCGCUCCUCUUGUCGGGCAUCGCCUCCUGGCCCUGGUCGCAGUA---UCGCCAGGUGCCCGACGAUGCCAGCGAACUAUUUAUCG ..---(((.(..(((((.......(((((.....))))).....((((((((((..(((....)))(((((.((.....---.))))))))))))))))).)))))..).)))....... ( -47.60) >DroAna_CAF1 861 117 + 1 AAUUUUAGAUAAUUGGCUACAUGUGCGCCUUGGAGGCAUUUCUUUUGUCGGGCAUCGCAUCCUGGCCGUGGACUCAGUA---CCGUCAGGUGCCCGACGAUGCCAGCGAAUUGUUUAUCG .....(((((((((.(((....(.(((((.....))).......((((((((((......((((((.(((.......))---).)))))))))))))))).)))))).)))))))))... ( -41.90) >consensus CA___UAGAUAAUUGGCUAUAUUUGCGCCUUGGAGGCACUUCUAUUGUCGGGCAUCGCCUCCUGGCCCUGGACUCAGUA___UCGCCAGGUGCCCGACGAUGCCAGCGAACUGUUUAUCG .....(((.(..(((((.........(((.....)))......((((((((((((((((....)))..(((..............)))))))))))))))))))))..).)))....... (-29.14 = -29.22 + 0.09)

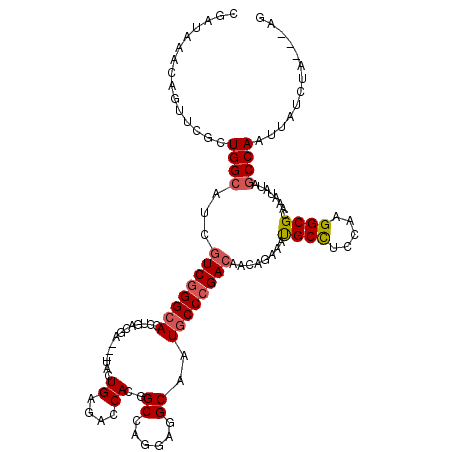
| Location | 18,849,915 – 18,850,029 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -23.78 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18849915 114 - 22407834 CGAUGAACAAUUCGCUGGCAUCGUCGGGCACCUGGCGG---UACUGAGUCCACGGCCAGGAUGCGAUGCCCGACAGUAGAAAUGCUUCCACGGCGCACAGAUAGCCAAUUAUCUA---UU .(((((........(((((..(((((((((((((((.(---(.........)).))))))......))))))))((((....)))).....)..)).)))........)))))..---.. ( -40.69) >DroVir_CAF1 951 114 - 1 CAAUGAACAGCUCGCUGGCAUCGUCGGGCACCUGGCGA---UACUGAGACCAGGGCCAGGAGGCAAUGCCCGACAGCAGCAGCGCCUCCAAGGCGCAAAUAAAGCCAAUUAGCUA---AG ........((((.(.((((...((((((((((((((..---......).)))))(((....)))..)))))))).......(((((.....))))).......)))).).)))).---.. ( -51.20) >DroGri_CAF1 1234 114 - 1 CGAUAAACAGUUCGCUGGCAUCGUCGGGCACCUGACGA---UAUUGGGACCAGGGCCACGAGGCAAUGCCGGACAACAGCAGUGCCUCCAAGGCGCAAAUAUAGCCAAUUAUCUA---AG .(((((.....(((.((((((((((((....)))))))---).(((....))).)))))))(((..(((.(.....).)))(((((.....))))).......)))..)))))..---.. ( -42.20) >DroWil_CAF1 836 117 - 1 CAAUAAAUAGUUCACUGGCAUCAUCGGGAACCUGACGAUAAUACUGAAUCCACGGCCACGAAGCAAUGCCCGACAAUAGAAAUGCCUCCAAGGCGCAAAGAUAACCAAUUAUCUA---UA .........(((((.((..(((.((((....)))).)))..)).)))))...(((.((........)).)))...........(((.....)))....((((((....)))))).---.. ( -20.90) >DroMoj_CAF1 629 114 - 1 CGAUAAAUAGUUCGCUGGCAUCGUCGGGCACCUGGCGA---UACUGCGACCAGGGCCAGGAGGCGAUGCCCGACAAGAGGAGCGCCUCCAAGGCGCAAAUGUAGCCAAUUAGCUA---AG .......(((((.(.((((...(((((((((((((((.---.....)).)))))(((....)))..)))))))).......(((((.....))))).......)))).).)))))---.. ( -51.90) >DroAna_CAF1 861 117 - 1 CGAUAAACAAUUCGCUGGCAUCGUCGGGCACCUGACGG---UACUGAGUCCACGGCCAGGAUGCGAUGCCCGACAAAAGAAAUGCCUCCAAGGCGCACAUGUAGCCAAUUAUCUAAAAUU (((........))).((((...((((((((((....))---).(((.((.....)))))........))))))).........(((.....))).........))))............. ( -33.70) >consensus CGAUAAACAGUUCGCUGGCAUCGUCGGGCACCUGACGA___UACUGAGACCACGGCCAGGAGGCAAUGCCCGACAACAGAAAUGCCUCCAAGGCGCAAAUAUAGCCAAUUAUCUA___AG ...............((((...((((((((..............((....))..((......))..))))))))........((((.....))))........))))............. (-23.78 = -24.08 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:07 2006