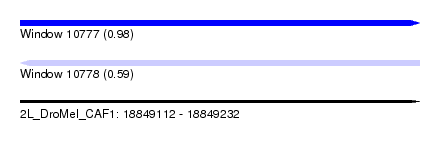
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,849,112 – 18,849,232 |
| Length | 120 |
| Max. P | 0.983938 |

| Location | 18,849,112 – 18,849,232 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.83 |
| Mean single sequence MFE | -53.28 |
| Consensus MFE | -43.79 |
| Energy contribution | -44.02 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
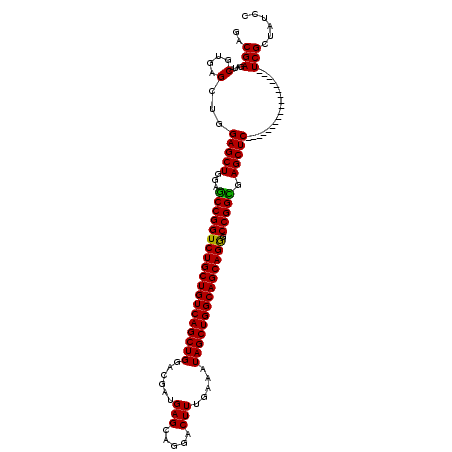
>2L_DroMel_CAF1 18849112 120 + 22407834 GACGAGAUUCGGGAGCUGGAGCUGGAGCCGGUCUGCUGUCAGCUGGACGAUGAGCAGGACUUUGAAAUAGCUGGCAGCAGGGCCGGCGAGCUCUCCCACUCGCACUCGCACUCGCUGUCC ((((((...((((.(..((((((...(((((((((((((((((((......(((.....))).....)))))))))))).))))))).))))))..).))))..)))((....)).))). ( -56.60) >DroVir_CAF1 73 102 + 1 GACGAGAUACGUGAGCUGGAGCUGGAGCCGGUCUGCUGUCAGCUGGACGAUGAGCAGGACUUUGAAAUAGCUGGCAGCAGGGCCGGCGAGCUC------------------UCGCUCUCA ..........(.((((.((((((...(((((((((((((((((((......(((.....))).....)))))))))))).))))))).)))))------------------).)))).). ( -54.20) >DroGri_CAF1 73 102 + 1 GACGAGAUACGUGAGCUGGAGCUGGAGCCGGUCUGCUGUCAGCUGGACGAUGAGCAGGACUUUGAAAUAGCUGGCAGCAGGGCCGGUGAGCUC------------------UCGCUCUCA ..........(.((((.((((((...(((((((((((((((((((......(((.....))).....)))))))))))).))))))).)))))------------------).)))).). ( -52.60) >DroWil_CAF1 73 102 + 1 GACGAGAUACGUGAGCUGGAGCUGGAACCGGUCUGCUGUCAGCUGGACGAUGAGCAGGACUUUGAAAUAGCUGGCAGCAGAGCCGGUGAGCUC------------------UCGCUAUCC .(((.....))).(((.((((((...(((((((((((((((((((......(((.....))).....)))))))))))))).))))).)))))------------------).))).... ( -49.50) >DroAna_CAF1 73 120 + 1 GACGAGAUACGGGAGUUGGAGCUGGAGCCGGUCUGCUGUCAGCUGGACGAUGAGCAGGACUUUGAGAUAGCUGGCAGCAGGGCCGGCGAGCUCUCCCACUCGCAUUCGCACUCGCUGUCC ..((((....((((....(((((...(((((((((((((((((((..(...(((.....)))...).)))))))))))).))))))).)))))))))....((....)).))))...... ( -56.00) >DroPer_CAF1 11851 102 + 1 GACGAGAUACGUGAGCUGGAGCUGGAGCCGGUCUGCUGUCAGCUGGACGAUGAGCAGGACUUUGAAAUAGCUGGCAGCAGGGCCGGCGAGCUC------------------UCGCUAUCC .(((.....))).(((.((((((...(((((((((((((((((((......(((.....))).....)))))))))))).))))))).)))))------------------).))).... ( -50.80) >consensus GACGAGAUACGUGAGCUGGAGCUGGAGCCGGUCUGCUGUCAGCUGGACGAUGAGCAGGACUUUGAAAUAGCUGGCAGCAGGGCCGGCGAGCUC__________________UCGCUAUCC .....((((.((((....(((((...(((((((((((((((((((......(((.....))).....)))))))))))))).))))).)))))..................)))))))). (-43.79 = -44.02 + 0.22)

| Location | 18,849,112 – 18,849,232 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.83 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -32.34 |
| Energy contribution | -32.34 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
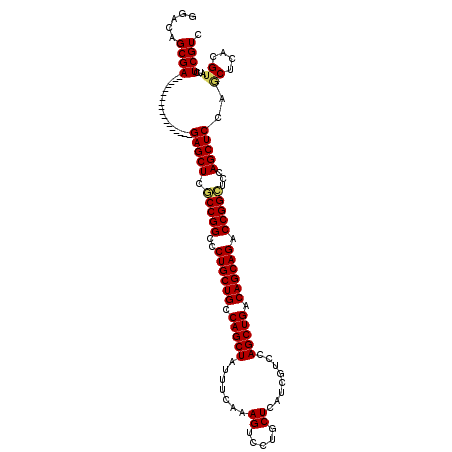
>2L_DroMel_CAF1 18849112 120 - 22407834 GGACAGCGAGUGCGAGUGCGAGUGGGAGAGCUCGCCGGCCCUGCUGCCAGCUAUUUCAAAGUCCUGCUCAUCGUCCAGCUGACAGCAGACCGGCUCCAGCUCCAGCUCCCGAAUCUCGUC .....(((((..((.(...((((.((((.....(((((..((((((.(((((.......((.....))........))))).)))))).))))).....)))).)))))))...))))). ( -45.46) >DroVir_CAF1 73 102 - 1 UGAGAGCGA------------------GAGCUCGCCGGCCCUGCUGCCAGCUAUUUCAAAGUCCUGCUCAUCGUCCAGCUGACAGCAGACCGGCUCCAGCUCCAGCUCACGUAUCUCGUC ...((((..------------------(((((.(((((..((((((.(((((.......((.....))........))))).)))))).)))))...)))))..))))(((.....))). ( -42.26) >DroGri_CAF1 73 102 - 1 UGAGAGCGA------------------GAGCUCACCGGCCCUGCUGCCAGCUAUUUCAAAGUCCUGCUCAUCGUCCAGCUGACAGCAGACCGGCUCCAGCUCCAGCUCACGUAUCUCGUC ...((((..------------------(((((..((((..((((((.(((((.......((.....))........))))).)))))).))))....)))))..))))(((.....))). ( -38.56) >DroWil_CAF1 73 102 - 1 GGAUAGCGA------------------GAGCUCACCGGCUCUGCUGCCAGCUAUUUCAAAGUCCUGCUCAUCGUCCAGCUGACAGCAGACCGGUUCCAGCUCCAGCUCACGUAUCUCGUC (((((((..------------------(((((.(((((.(((((((.(((((.......((.....))........))))).))))))))))))...)))))..))).....)))).... ( -38.36) >DroAna_CAF1 73 120 - 1 GGACAGCGAGUGCGAAUGCGAGUGGGAGAGCUCGCCGGCCCUGCUGCCAGCUAUCUCAAAGUCCUGCUCAUCGUCCAGCUGACAGCAGACCGGCUCCAGCUCCAACUCCCGUAUCUCGUC .....(((((((((...(.((((.((((.....(((((..((((((.(((((.......((.....))........))))).)))))).))))).....)))).))))))))).))))). ( -47.56) >DroPer_CAF1 11851 102 - 1 GGAUAGCGA------------------GAGCUCGCCGGCCCUGCUGCCAGCUAUUUCAAAGUCCUGCUCAUCGUCCAGCUGACAGCAGACCGGCUCCAGCUCCAGCUCACGUAUCUCGUC (((((((..------------------(((((.(((((..((((((.(((((.......((.....))........))))).)))))).)))))...)))))..))).....)))).... ( -37.76) >consensus GGACAGCGA__________________GAGCUCGCCGGCCCUGCUGCCAGCUAUUUCAAAGUCCUGCUCAUCGUCCAGCUGACAGCAGACCGGCUCCAGCUCCAGCUCACGUAUCUCGUC .....((((..................(((((.(((((..((((((.(((((.......((.....))........))))).)))))).)))))...)))))..((....))...)))). (-32.34 = -32.34 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:04 2006