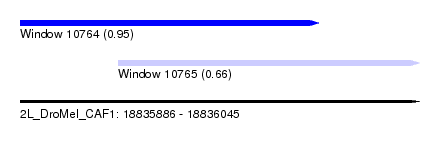
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,835,886 – 18,836,045 |
| Length | 159 |
| Max. P | 0.948356 |

| Location | 18,835,886 – 18,836,005 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -32.06 |
| Energy contribution | -31.73 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
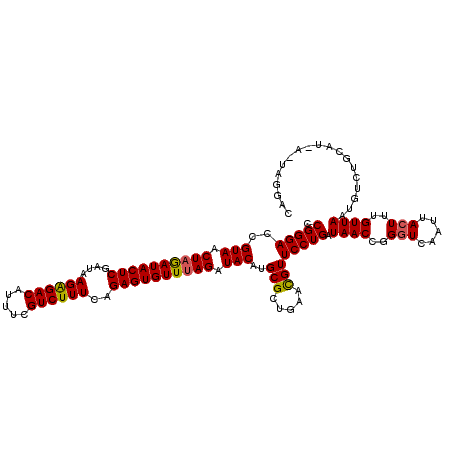
>2L_DroMel_CAF1 18835886 119 + 22407834 UUUCAGCGAGUGGACGCUU-AAAAGCUUUUUUAAUUAAUUCCGGGACCGUAACUAGAUACUCCAUGAGGGACAUUUCGUCUUUCAGAGUGUUCAGAUACAUGCGCUGAAUGUUCCUGAUA .(((((((.(((...(((.-...)))...............((....))...((..((((((..((((((((.....))))))))))))))..))...))).)))))))........... ( -32.80) >DroSec_CAF1 1801 120 + 1 UUUCAGCGAGUGGAUGCUCUAUAAGCUUUUUUAAUUAAUUCCGGGACCGUAACUAGAUACUCGAUAAGAGACAUUUCGUCUUUCAGAGUGUUUAGAUACAUGCGCUGAACGUUCCUGAUA .((((((((((....)))).....((................(....)(((.((((((((((....((((((.....))))))..)))))))))).)))..))))))))........... ( -34.00) >DroSim_CAF1 42 119 + 1 UUUCAGCGAGUGGACGCUC-AUAAGCUUUUUUAAUUAAUUCCGGGACCGUAACUAAAUACUCGAUAAGAGACAUUUCGUCUUUCAGAGUGUUUAGAUACAUGCGCUGAACGUUCCUGAUA .((((((((((....))))-....((................(....)(((.((((((((((....((((((.....))))))..)))))))))).)))..))))))))........... ( -34.90) >consensus UUUCAGCGAGUGGACGCUC_AUAAGCUUUUUUAAUUAAUUCCGGGACCGUAACUAGAUACUCGAUAAGAGACAUUUCGUCUUUCAGAGUGUUUAGAUACAUGCGCUGAACGUUCCUGAUA .((((((((((....)))).....((................(....)(((.((((((((((....((((((.....))))))..)))))))))).)))..))))))))........... (-32.06 = -31.73 + -0.33)



| Location | 18,835,925 – 18,836,045 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -27.09 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18835925 120 + 22407834 CCGGGACCGUAACUAGAUACUCCAUGAGGGACAUUUCGUCUUUCAGAGUGUUCAGAUACAUGCGCUGAAUGUUCCUGAUAACCGGAUCAAUUACUUUGUUAAUAUCUGCAUCAGUAGGAC ((((............((((((..((((((((.....))))))))))))))((((..((((.......))))..))))...))))...................(((((....))))).. ( -31.70) >DroSec_CAF1 1841 105 + 1 CCGGGACCGUAACUAGAUACUCGAUAAGAGACAUUUCGUCUUUCAGAGUGUUUAGAUACAUGCGCUGAACGUUCCUGAUAACCGGGUCAAUUACUUUGUUAAUGU--------------- .(((((..(((.((((((((((....((((((.....))))))..)))))))))).)))..(((.....)))))))).((((..(((.....)))..))))....--------------- ( -30.50) >DroSim_CAF1 81 120 + 1 CCGGGACCGUAACUAAAUACUCGAUAAGAGACAUUUCGUCUUUCAGAGUGUUUAGAUACAUGCGCUGAACGUUCCUGAUAACCGGGUCAAUUACUUUGUUAAUGUCUGCAUAACUAGGAC ((((....(((.((((((((((....((((((.....))))))..)))))))))).)))..(((.....))).........))))........(((.((((.((....)))))).))).. ( -31.90) >consensus CCGGGACCGUAACUAGAUACUCGAUAAGAGACAUUUCGUCUUUCAGAGUGUUUAGAUACAUGCGCUGAACGUUCCUGAUAACCGGGUCAAUUACUUUGUUAAUGUCUGCAU_A_UAGGAC .(((((..(((.((((((((((....((((((.....))))))..)))))))))).)))..(((.....)))))))).((((..(((.....)))..))))................... (-27.09 = -27.10 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:52 2006