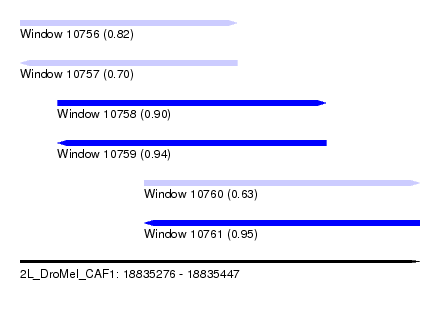
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,835,276 – 18,835,447 |
| Length | 171 |
| Max. P | 0.952355 |

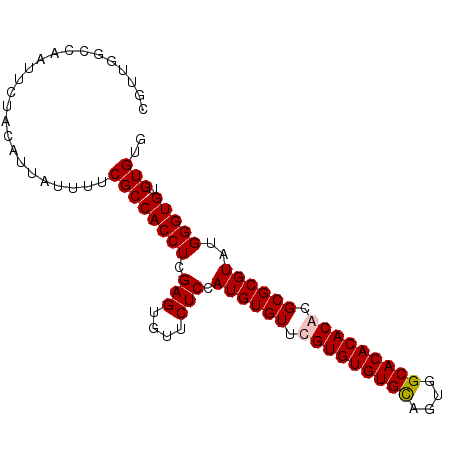
| Location | 18,835,276 – 18,835,369 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18835276 93 + 22407834 CGUUGGCCAAUUCUACAUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUUGUGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUG ........................((((((((.(((....))).((((((.(((((((((....))))))))).))))))..))))).))).. ( -32.70) >DroSim_CAF1 2627 93 + 1 CGUUGGCCAAUUCUACAUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUUGUGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUG ........................((((((((.(((....))).((((((.(((((((((....))))))))).))))))..))))).))).. ( -32.70) >DroEre_CAF1 2698 93 + 1 CGUUCGCCAAUUCUACGUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUCGUGUGUGUAGUGGCACACACACGCGCGUAUGGGUGUGUGUG ........................(((((((..(((....)))(((((((.(((((((((......))))))))).))))))))))).))).. ( -29.50) >DroYak_CAF1 2729 93 + 1 CGUUGGCCAAUUCUACGUUAUUUUCGCCACCUCGAGUAUUCUCCAUGUGUUCGUGUGUGUAGUGGCACACACACGCGCGUAGGGGUGUGUGUG (((.((.....)).))).......((((((((((((....))).((((((..((((((((....))))))))..)))))).)))))).))).. ( -32.00) >consensus CGUUGGCCAAUUCUACAUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUCGUGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUG ........................((((((((.(((....))).((((((.(((((((((....))))))))).))))))..))))).))).. (-30.60 = -30.85 + 0.25)

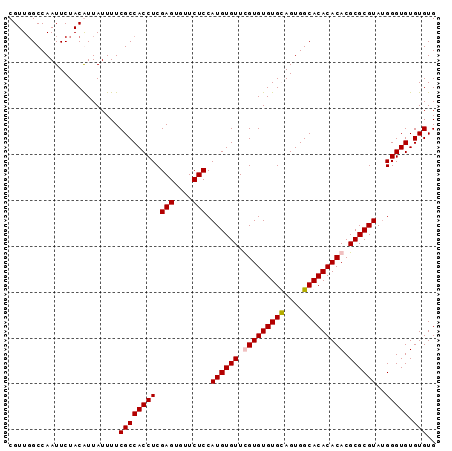
| Location | 18,835,276 – 18,835,369 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.83 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18835276 93 - 22407834 CACACACACCCAUACGCGCGUGUGUGUGCCACUGCACACACAAACACAUGGAGAACACUCGAGGUGGCGAAAAUAAUGUAGAAUUGGCCAACG .((((((((.(......).))))))))(((((((((........(((.(.(((....))).).)))..........)))))...))))..... ( -25.27) >DroSim_CAF1 2627 93 - 1 CACACACACCCAUACGCGCGUGUGUGUGCCACUGCACACACAAACACAUGGAGAACACUCGAGGUGGCGAAAAUAAUGUAGAAUUGGCCAACG .((((((((.(......).))))))))(((((((((........(((.(.(((....))).).)))..........)))))...))))..... ( -25.27) >DroEre_CAF1 2698 93 - 1 CACACACACCCAUACGCGCGUGUGUGUGCCACUACACACACGAACACAUGGAGAACACUCGAGGUGGCGAAAAUAACGUAGAAUUGGCGAACG ..............(((.((((((((((....))))))))))..(((.(.(((....))).).)))))).......(((.......))).... ( -26.10) >DroYak_CAF1 2729 93 - 1 CACACACACCCCUACGCGCGUGUGUGUGCCACUACACACACGAACACAUGGAGAAUACUCGAGGUGGCGAAAAUAACGUAGAAUUGGCCAACG ...........(((((((((((((((((....)))))))))...(((.(.(((....))).).)))))).......)))))............ ( -26.41) >consensus CACACACACCCAUACGCGCGUGUGUGUGCCACUACACACACAAACACAUGGAGAACACUCGAGGUGGCGAAAAUAACGUAGAAUUGGCCAACG .((((((((.(......).))))))))((((((((.........(((.(.(((....))).).)))...........))))...))))..... (-24.08 = -23.83 + -0.25)

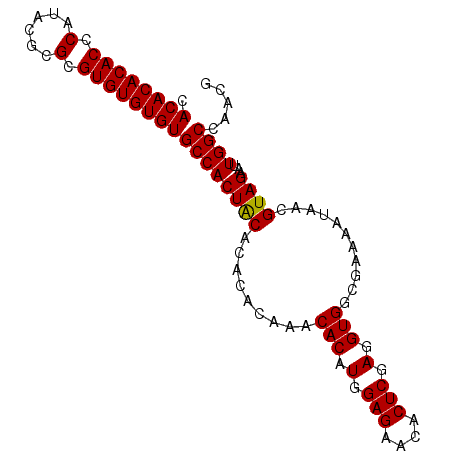
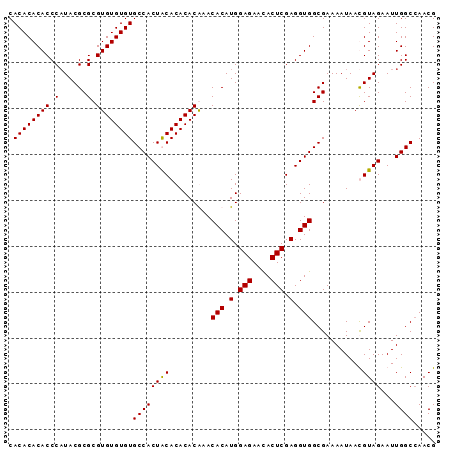
| Location | 18,835,292 – 18,835,407 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -40.01 |
| Energy contribution | -40.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18835292 115 + 22407834 AUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUUGUGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGU--GUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCG ........((((((((.(((....))).((((((.(((((((((....))))))))).))))))..))))).))).((((--(......)))))(((.((((.....)))))))... ( -41.40) >DroSim_CAF1 2643 117 + 1 AUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUUGUGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCG ...............((((.(((((.....((((.(((((((((....))))))))).))))(((((((((((.(.(((.....)))).))))))))))).)))))))))....... ( -42.50) >DroEre_CAF1 2714 117 + 1 GUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUCGUGUGUGUAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCG ...............((((.(((((..((((....)))).((((((.((((.((((((((((((((....)))))))))))))))))))))))).......)))))))))....... ( -42.50) >DroYak_CAF1 2745 117 + 1 GUUAUUUUCGCCACCUCGAGUAUUCUCCAUGUGUUCGUGUGUGUAGUGGCACACACACGCGCGUAGGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCG ...............((((.(((((..((((....)))).((((((.((((.(((((((((((((......))))))))))))))))))))))).......)))))))))....... ( -40.80) >consensus AUUAUUUUCGCCACCUCGAGUGUUCUCCAUGUGUUCGUGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCG ...............((((.(((((.....((((.(((((((((....))))))))).))))(((((((((((.(.(((.....)))).))))))))))).)))))))))....... (-40.01 = -40.33 + 0.31)


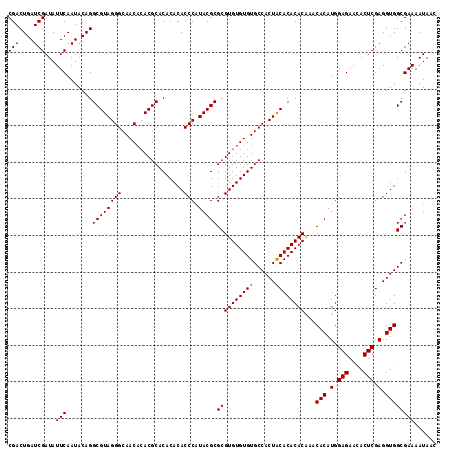
| Location | 18,835,292 – 18,835,407 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -32.68 |
| Energy contribution | -32.43 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18835292 115 - 22407834 CGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAAC--ACGCACACACACCCAUACGCGCGUGUGUGUGCCACUGCACACACAAACACAUGGAGAACACUCGAGGUGGCGAAAAUAAU (((....)))...(((.......((((((((....--...........))).)))))((.(((((((((....)))))))))..(((.(.(((....))).).))))))))...... ( -34.36) >DroSim_CAF1 2643 117 - 1 CGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCAUACGCGCGUGUGUGUGCCACUGCACACACAAACACAUGGAGAACACUCGAGGUGGCGAAAAUAAU (((....)))...(((.......((((((((.................))).)))))((.(((((((((....)))))))))..(((.(.(((....))).).))))))))...... ( -34.23) >DroEre_CAF1 2714 117 - 1 CGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCAUACGCGCGUGUGUGUGCCACUACACACACGAACACAUGGAGAACACUCGAGGUGGCGAAAAUAAC ...(((((.(.....).)).)))((((((((((((((((((.(............).)))))))).)))).)))).........(((.(.(((....))).).)))))......... ( -34.10) >DroYak_CAF1 2745 117 - 1 CGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCCUACGCGCGUGUGUGUGCCACUACACACACGAACACAUGGAGAAUACUCGAGGUGGCGAAAAUAAC ((.(((.(((((((((.......((((((((..................)))))))).((((((((((....))))))))))..........))))).))))..)))))........ ( -38.17) >consensus CGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCAUACGCGCGUGUGUGUGCCACUACACACACAAACACAUGGAGAACACUCGAGGUGGCGAAAAUAAC (((....)))...(((.......((((((((.................))).)))))((((((((((......))))))))...(((.(.(((....))).).))))))))...... (-32.68 = -32.43 + -0.25)

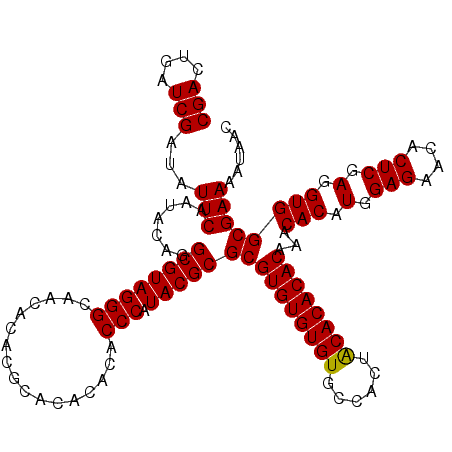
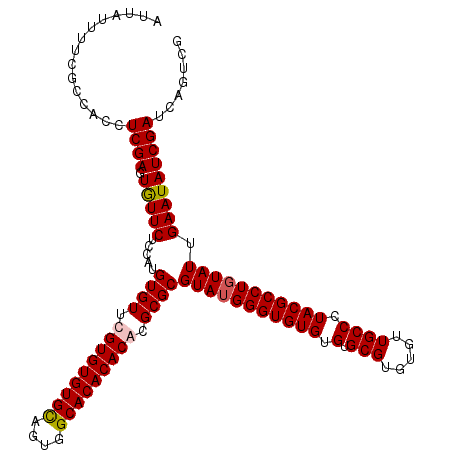
| Location | 18,835,329 – 18,835,447 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -45.22 |
| Consensus MFE | -42.38 |
| Energy contribution | -43.62 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18835329 118 + 22407834 UGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGU--GUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCGGCCGUCUACGUGACGCCUCUCAGCUAGCCGUGUCUUUUAU .(((((..(..((((.(((((((((......))))))))).))--))..)..)))))(((.((((.....))))))).((((.(((.....)))((......))..)))).......... ( -43.70) >DroSim_CAF1 2680 120 + 1 UGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCGGCCGUCUACGUGACGCCUCUCAGCUAGCCGUGUCUUUUAU .(((((..(..((((((.((((((((......)))))))).))))))..)..)))))(((.((((.....))))))).((((.(((.....)))((......))..)))).......... ( -43.90) >DroEre_CAF1 2751 120 + 1 UGUGUGUAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCGGCCGUCUACGUGACGCCUCUCAGCUAGCCGUGUCUUUUAU ...((((((.((((.((((((((((((((....))))))))))))))))))))))))(((.((((.....))))))).((((.(((.....)))((......))..)))).......... ( -47.50) >DroYak_CAF1 2782 120 + 1 UGUGUGUAGUGGCACACACACGCGCGUAGGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCGGCCGUCUACGUGACGCCUCUCAGCUAGCCGUGUCUUUUAU ...((((((.((((.(((((((((((((......)))))))))))))))))))))))(((.((((.....))))))).((((.(((.....)))((......))..)))).......... ( -45.80) >consensus UGUGUGCAGUGGCACACACACGCGCGUAUGGGUGUGUGUGCGUGUGUUGCCCUACGCCUGUAUUGAAUAUCGAUCAGUCGGCCGUCUACGUGACGCCUCUCAGCUAGCCGUGUCUUUUAU ...((((((.((((.((((((((((((((....))))))))))))))))))))))))(((.((((.....))))))).((((.(((.....)))((......))..)))).......... (-42.38 = -43.62 + 1.25)


| Location | 18,835,329 – 18,835,447 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -40.45 |
| Energy contribution | -40.70 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18835329 118 - 22407834 AUAAAAGACACGGCUAGCUGAGAGGCGUCACGUAGACGGCCGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAAC--ACGCACACACACCCAUACGCGCGUGUGUGUGCCACUGCACACA ..........(((((.(((....)))(((.....)))))))).(((((.(.....).)).)))(.((((((((.(--((((((.(.(........).).))))))))))).)))).)... ( -41.00) >DroSim_CAF1 2680 120 - 1 AUAAAAGACACGGCUAGCUGAGAGGCGUCACGUAGACGGCCGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCAUACGCGCGUGUGUGUGCCACUGCACACA ..........(((((.(((....)))(((.....)))))))).(((((.(.....).)).)))(.((((((((((((((((.(............).)))))))).)))).)))).)... ( -42.30) >DroEre_CAF1 2751 120 - 1 AUAAAAGACACGGCUAGCUGAGAGGCGUCACGUAGACGGCCGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCAUACGCGCGUGUGUGUGCCACUACACACA ..........(((((.(((....)))(((.....)))))))).(((((.(.....).)).)))(.((((((((((((((((.(............).)))))))).)))).)))).)... ( -42.30) >DroYak_CAF1 2782 120 - 1 AUAAAAGACACGGCUAGCUGAGAGGCGUCACGUAGACGGCCGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCCUACGCGCGUGUGUGUGCCACUACACACA ..........(((((.(((....)))(((.....)))))))).(((((.(.....).)).)))(.((((((((((((((((.(............).)))))))).)))).)))).)... ( -42.30) >consensus AUAAAAGACACGGCUAGCUGAGAGGCGUCACGUAGACGGCCGACUGAUCGAUAUUCAAUACAGGCGUAGGGCAACACACGCACACACACCCAUACGCGCGUGUGUGUGCCACUACACACA ..........(((((.(((....)))(((.....)))))))).(((((.(.....).)).)))(.((((((((((((((((.(............).)))))))).)))).)))).)... (-40.45 = -40.70 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:49 2006