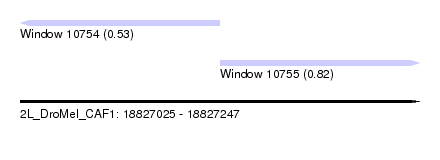
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,827,025 – 18,827,247 |
| Length | 222 |
| Max. P | 0.817411 |

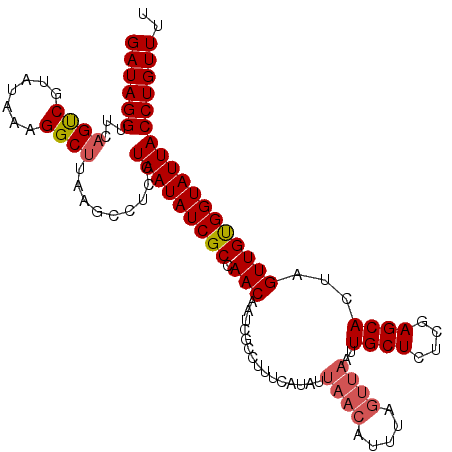
| Location | 18,827,025 – 18,827,136 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18827025 111 - 22407834 GGGCUAUCGAUACCGAUUUCCACAGGGUGACCACAUUGCCAUAACAAAAAAUUUAGGG------AAGCGAAUAUUCAAGU-ACGCGCCCUGCGUA-CAUA-UGUGCAGCCCUGAAUAAAG .(((.((((....)))).......((....)).....)))..........((((((((------..(((.((((....((-((((.....)))))-))))-).)))..)))))))).... ( -34.20) >DroSec_CAF1 95818 118 - 1 GGGCUAUCGAUACCGAUUUCCACAGGGUGACCACAUUGCUUCAACAAAAAUUUGAGGGUACACUAAGCGAAUGUUCAAGU-ACGCGCCCUGCGAA-CAUAGUGUGCAGCCCUGGGUAAAG .((..((((....))))..)).((((((..........((((((.......))))))((((((((...((....))..((-.(((.....))).)-).)))))))).))))))....... ( -35.10) >DroSim_CAF1 92053 118 - 1 GGGCUAUCGAUACCGAUUUCCACAGGGUGACCACAUUGCUUCAACAAAAAUUUGAGGGUACACUAAACGAAUGUUCAAGU-ACGCGCCCUGCGAA-CAUAGUGUGCAGCCCUGGGUAAAG .((..((((....))))..)).((((((..........((((((.......))))))((((((((...((....))..((-.(((.....))).)-).)))))))).))))))....... ( -35.10) >DroEre_CAF1 90862 119 - 1 GGGCUAUCGAUACCGGUAUGCACAGGGUGACCAAACUGCGUUAG-AGUAACAAGCGGUUACACUACGCGAAUGUUCAAGUUGCGCGCCUUGACUACAAUAGUGUGCAGCCCUGGAAAAAA (((((((((....)))))((((((((....))....(((((.((-.(((((.....))))).)))))))..((((((((........)))))..)))....))))))))))......... ( -39.10) >DroYak_CAF1 91680 97 - 1 G-CUUAUCGAUACCGAAAUGCACAGGGUGACCAGAUUUCGUUUG-AAC-----------ACAC----------UUCAAGUGGCGCCCGCUUCAUAAAAUAGUGUGCAGCCCUGGAAAAAU (-(...(((....)))...)).((((((...(((((...)))))-..(-----------((((----------(..((((((...))))))........))))))..))))))....... ( -25.30) >consensus GGGCUAUCGAUACCGAUUUCCACAGGGUGACCACAUUGCGUUAACAAAAAUUUGAGGGUACACUAAGCGAAUGUUCAAGU_ACGCGCCCUGCGUA_CAUAGUGUGCAGCCCUGGAUAAAG (((((((((....)))).......((....))...................................................((((.(((.......))).)))))))))......... (-16.16 = -16.92 + 0.76)

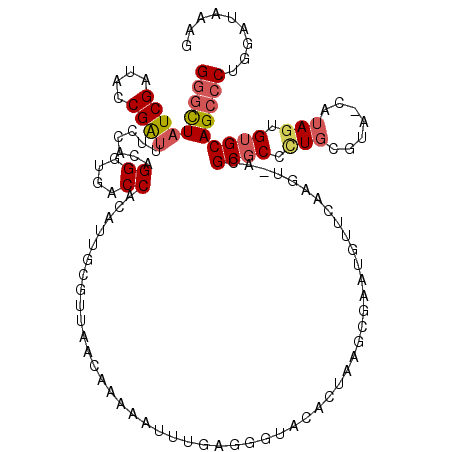
| Location | 18,827,136 – 18,827,247 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -20.66 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18827136 111 + 22407834 GAUAGGUUCAGUCGUAUGAAGGCUUAAGCCUCCUAAUAUCGCCAACAAUCGCCAUUCAUUUUAACAUUUAGUUAAUUGCUCUUGAGCACUAGUUGUGGUAUUACCUGUUUU ((((((((((......))))(((....)))...(((((((((.(((..............(((((.....))))).((((....))))...)))))))))))))))))).. ( -25.70) >DroSec_CAF1 95936 103 + 1 GAUAGGUUCAGCCGUAUAAAGGCUUAAGCCUCCUAAUAUCGCAAACAAUCGCCUUUCAUAUUAACAUU--------UGCUCUCGAGCACUAGUUGCGGUAUUACCUGUUUU (((((((...((((((...((((....)))).(((.....((........))................--------((((....)))).))).))))))...))))))).. ( -24.30) >DroSim_CAF1 92171 111 + 1 GAUAGGUUCAGUCGUAUAAAGGCUUAAGCCUCCUAAUAUCGCCAACAAUCGCCUUUCAUAUUAACAUUUAGUUAAUUGCUCUCGAGCACUAGUUGCGGUAUUACCUGUUUU ((((((.............((((....))))..(((((((((.(((.............((((((.....))))))((((....))))...)))))))))))))))))).. ( -27.60) >DroEre_CAF1 90981 111 + 1 GAUAGGUUCAGUCGAAUAAAGGCUUAAGCCUCCUAAUAUCGCCAACAAUCGCAGUUCAUACUAACAUUUAGUUAAUUGCUCUCGAGCACUAGUUGUGGUAUUACCUGUUUU ((((((.............((((....))))..(((((((((.(((...............((((.....))))..((((....))))...)))))))))))))))))).. ( -24.30) >DroYak_CAF1 91777 104 + 1 GAUAGGUUCAGUCGGAUAAAGGC-------UCCUAAUAUCGCCAACAAUCGCCUUUCAUACUAACAUUUAGUUAAUUGCUCUCGAGCAUUAGUUGUGGUAUUACCUGUUUU ((((((...((((.......)))-------)..(((((((((.(((...............((((.....))))..((((....))))...)))))))))))))))))).. ( -21.90) >consensus GAUAGGUUCAGUCGUAUAAAGGCUUAAGCCUCCUAAUAUCGCCAACAAUCGCCUUUCAUAUUAACAUUUAGUUAAUUGCUCUCGAGCACUAGUUGUGGUAUUACCUGUUUU ((((((...((((.......)))).........(((((((((.(((...............((((.....))))..((((....))))...)))))))))))))))))).. (-20.66 = -21.26 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:44 2006