
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,816,633 – 18,816,793 |
| Length | 160 |
| Max. P | 0.904866 |

| Location | 18,816,633 – 18,816,753 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -22.02 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
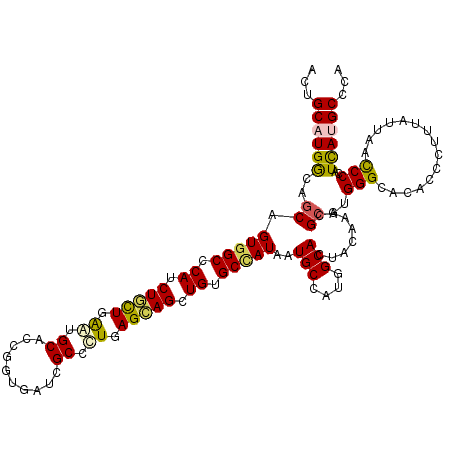
>2L_DroMel_CAF1 18816633 120 + 22407834 AUUCGAUAUGACGUCAUACUCCUGGAACACUUCUCCAACGAUUGCAUGGCAGCGGUGGCCCACCUAUUGAAUGCACCUGUAAUCGCAUUGAGUAGCUGUGCGAUAAUGCCGUGGCACUAC ..(((.((((....))))....((((.......)))).))).(((((((((...((.((.((.(((((.(((((..........))))).))))).)).)).))..)))))).))).... ( -34.90) >DroVir_CAF1 126871 120 + 1 CAGCGCUACGAUGUCAUCAUAUUGGAGCACUUCUCCAAUGAUUGCCUGGCCGCGGUGGCCCAUCUGCUGGAUGCCCCGGUCAUCGCCUUGAGCAGCUGUGCCAUUAUGCCCUGGCAUUAU (((((((.(((.((((.((((((((((.....))))))))..))..)))).(((((((((((((.....))))....))))))))).)))))).))))(((((........))))).... ( -42.50) >DroPse_CAF1 94293 120 + 1 CUGCGCUACGAUCUGAUCAUCCUGGAGCAUUUCGCCAAUGACUGCAUGACCGCAGUGGCGCAUCUGCUGAACGCACCGGUGAUCGCUCUCAGCAGCUGUGCCAUCAUGCCAUGGCACUAC .(((.(((.(((......))).))).)))....((((.(((((((......)))))((((((.(((((((..((..........))..))))))).)))))).......))))))..... ( -43.50) >DroGri_CAF1 110046 120 + 1 CGGCGAUAUGAUGUCAUCAUUCUGGAGCACUUCUCCAACGAUUGCUUGGCAGCUGUGGCACAUCUACUAGAUGCACCCGUCAUAGCGUUAAGUAGUUGUGCCAUUAUGCCGUGGCAUUAU .......((((((((((((.(((((((.....)))))..)).))...((((...((((((((.(((((.(((((..........))))).))))).))))))))..)))))))))))))) ( -47.10) >DroAna_CAF1 74474 120 + 1 AUCCGAUUCGAUGUGGUCAUUCUUGAGCACUUUUCCAACGACUGCAUGGCAGCUGUGGCCCACCUCUUGGGGGCUCCCAUGAUCGCCCUGAGUAGCUGUGCUAUAAUGCCAUGGCACUAC ...((.((.((.(((.(((....))).)))...)).))))..(((((((((..((((((.((.((((..((((.((....)).).)))..)).)).)).)))))).)))))).))).... ( -39.80) >DroPer_CAF1 94789 120 + 1 CUGCGCUACGAUCUGAUCAUCCUGGAGCACUUUGCCAAUGACUGCAUGACCGCAGUGGCGCAUCUGCUGAAUGCACCGGUGAUCGCUCUCAGCAGCUGUGCCAUCAUGCCCUGGCACUAU .(((.(((.(((......))).))).)))...(((((...(((((......)))))((((((.(((((((..((..........))..))))))).)))))).........))))).... ( -44.20) >consensus CUGCGAUACGAUGUCAUCAUCCUGGAGCACUUCUCCAACGACUGCAUGGCAGCAGUGGCCCAUCUGCUGAAUGCACCGGUGAUCGCCCUGAGCAGCUGUGCCAUAAUGCCAUGGCACUAC ......................(((((.....))))).....((((((((....(((((.((.(((((.((.((..........)).)).))))).)).)))))...))))).))).... (-22.02 = -23.00 + 0.98)


| Location | 18,816,673 – 18,816,793 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -41.11 |
| Consensus MFE | -26.33 |
| Energy contribution | -25.81 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18816673 120 + 22407834 AUUGCAUGGCAGCGGUGGCCCACCUAUUGAAUGCACCUGUAAUCGCAUUGAGUAGCUGUGCGAUAAUGCCGUGGCACUACAAACGCAUGGGAAGUCCGUUCAUUAACCCCAUUAUGCCCA ..(((((((((...((.((.((.(((((.(((((..........))))).))))).)).)).))..)))))).)))........(((((((................))))...)))... ( -34.29) >DroVir_CAF1 126911 120 + 1 AUUGCCUGGCCGCGGUGGCCCAUCUGCUGGAUGCCCCGGUCAUCGCCUUGAGCAGCUGUGCCAUUAUGCCCUGGCAUUAUAAGCGCAUGGGCACGCCCUUUAUUAAUCCCAUCAUGCCCA ..((((.((.((..(((((.((.(((((((.....))(((....)))...))))).)).)))))..)).)).))))...........((((((.....................)))))) ( -39.20) >DroPse_CAF1 94333 120 + 1 ACUGCAUGACCGCAGUGGCGCAUCUGCUGAACGCACCGGUGAUCGCUCUCAGCAGCUGUGCCAUCAUGCCAUGGCACUACAAGCGCAUGGGCACACCCCAUAUCAACCCGAUCAUGCCCA ...((((((.(((.((((((((.(((((((..((..........))..))))))).))))))))..(((....)))......))).(((((.....)))))..........))))))... ( -45.60) >DroGri_CAF1 110086 120 + 1 AUUGCUUGGCAGCUGUGGCACAUCUACUAGAUGCACCCGUCAUAGCGUUAAGUAGUUGUGCCAUUAUGCCGUGGCAUUAUAAACGCAUGGGAACACCAUUUAUUAACCCAAUUACGCCCA ..(((((((((...((((((((.(((((.(((((..........))))).))))).))))))))..))))).))))...........((((...............)))).......... ( -40.46) >DroAna_CAF1 74514 120 + 1 ACUGCAUGGCAGCUGUGGCCCACCUCUUGGGGGCUCCCAUGAUCGCCCUGAGUAGCUGUGCUAUAAUGCCAUGGCACUACAAGCGGAUGGGCAGUCCGUUCAUAAACCCCAUCAUGCCCA ..(((((((((..((((((.((.((((..((((.((....)).).)))..)).)).)).)))))).)))))).)))......(((((((((................)))))).)))... ( -42.09) >DroPer_CAF1 94829 120 + 1 ACUGCAUGACCGCAGUGGCGCAUCUGCUGAAUGCACCGGUGAUCGCUCUCAGCAGCUGUGCCAUCAUGCCCUGGCACUAUAAGCGCAUGGGCACACCCCAUAUCAACCCGAUUAUGCCCA ...((((((.((..((((((((.(((((((..((..........))..))))))).))))))))..(((((((((.......)).)).)))))...............)).))))))... ( -45.00) >consensus ACUGCAUGGCAGCAGUGGCCCAUCUGCUGAAUGCACCGGUGAUCGCCCUGAGCAGCUGUGCCAUAAUGCCAUGGCACUACAAGCGCAUGGGCACACCCUUUAUUAACCCCAUCAUGCCCA ...((((((..((.(((((.((.(((((.((.((..........)).)).))))).)).)))))..(((....)))........))..(((...............)))..))))))... (-26.33 = -25.81 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:40 2006