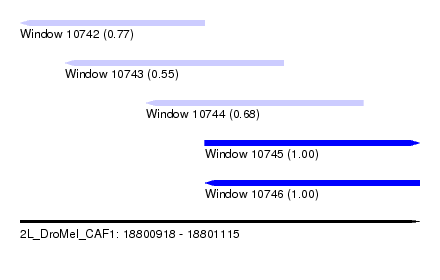
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,800,918 – 18,801,115 |
| Length | 197 |
| Max. P | 0.999570 |

| Location | 18,800,918 – 18,801,009 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -12.17 |
| Energy contribution | -12.37 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
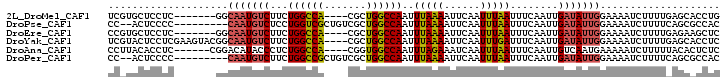
>2L_DroMel_CAF1 18800918 91 - 22407834 UCGUGCUCCUC-------GGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAUUGGAAAAUCUUUUGAGCACCUG ..((((((...-------..(((((((...(((((.----....)))))..(((((......)))))........)))))))(.....)....))))))... ( -22.10) >DroPse_CAF1 78839 91 - 1 CC--ACUCCCC---------CAAUGUCUCCUGGUCGCUGUCGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAUUGGAAAAUCUUUUCAGCGCCAC ..--......(---------(((((((...(((((((....)).)))))..(((((......)))))........))))))))................... ( -17.00) >DroEre_CAF1 64214 91 - 1 CCGUGCUCCUC-------GGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAUUGGAAAAUCUUUUGAGAAGCUC ....(((.(((-------(((((((((...(((((.----....)))))..(((((......)))))........)))))))(.....)..))))).))).. ( -20.20) >DroYak_CAF1 64003 98 - 1 UCGUACUCCUCGAAGUACGGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUGAUUUCAAUUGAUAUUGGAAAAUCUUUUGAGCACCUC .((((((......))))))((.........(((((.----....))))).........((((...(((((((((....))))))..)))..))))))..... ( -23.60) >DroAna_CAF1 58820 92 - 1 CCUUACACCUC------CGGACAUACCCUCUGGCCA----CGGUGGCCAAUUUAGAAAUCAAUUUAAUUUCAAUUGUCAAUGAAAAAUCUUUUUACACUCUC ...........------..((((.......(((((.----....))))).....(((((.......)))))...))))........................ ( -14.30) >DroPer_CAF1 78173 91 - 1 CC--ACUCCCC---------CAAUGUCUUCUGGCCGCUGUCGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAUUGGAAAAUCUUUUCAGCGCCAC ..--......(---------(((((((...(((((((....)).)))))..(((((......)))))........))))))))................... ( -19.70) >consensus CCGUACUCCUC_______GGCAAUGUCUUCUGGCCA____CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAUUGGAAAAUCUUUUGAGCACCUC ....................(((((((...((((((.......))))))..(((((......)))))........))))))).................... (-12.17 = -12.37 + 0.20)

| Location | 18,800,940 – 18,801,048 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
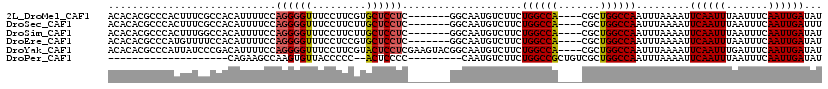
>2L_DroMel_CAF1 18800940 108 - 22407834 ACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCUUCGUGCUCCUC-------GGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAU ...................(((((.((.((((((.........)))))).-------)).)))))....(((((.----....))))).........((((((.......))))))... ( -21.40) >DroSec_CAF1 63448 108 - 1 ACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCUUCUUGCUCCUC-------GGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUUU ...................(((((.((.((((((.........)))))).-------)).)))))....(((((.----....))))).........((((((.......))))))... ( -21.40) >DroSim_CAF1 64075 108 - 1 ACACACGCCCACUUUGGCCACAUUUUCCAGGGGUUUCCUUCUUGCUCCUC-------GGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAU ......(((.....((((((((((.((.((((((.........)))))).-------)).))))).....)))))----....)))...........((((((.......))))))... ( -24.60) >DroEre_CAF1 64236 108 - 1 ACACACGCCCAUGUUUUCCACAUUUUCCAGGGGUUUCCUCCGUGCUCCUC-------GGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAU ......(((.((((.....)))).....((((((.........)))))).-------))).........(((((.----....))))).........((((((.......))))))... ( -21.40) >DroYak_CAF1 64025 115 - 1 ACACACGCCCAUUAUCCCGACAUUUUCCAGGGGUUUCCUUCGUACUCCUCGAAGUACGGCAAUGUCUUCUGGCCA----CGCUGGCCAAUUUAAAAUUCAAUUUGAUUUCAAUUGAUAU ......(((....(((((...........))))).......(((((......)))))))).........(((((.----....))))).........((((((.(....)))))))... ( -25.90) >DroPer_CAF1 78195 88 - 1 --------------------CAGAAGCCAAGUGUUACCCCC--ACUCCCC---------CAAUGUCUUCUGGCCGCUGUCGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAU --------------------.(((((.((((((.......)--)))....---------...)).)))))((((((....)).))))..........((((((.......))))))... ( -17.01) >consensus ACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCUUCGUGCUCCUC_______GGCAAUGUCUUCUGGCCA____CGCUGGCCAAUUUAAAAUUCAAUUUAAUUUCAAUUGAUAU ............................((((((.........))))))....................((((((.......)))))).........((((((.......))))))... (-14.38 = -14.52 + 0.14)
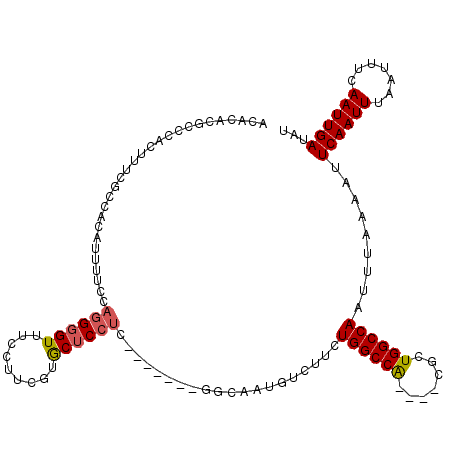
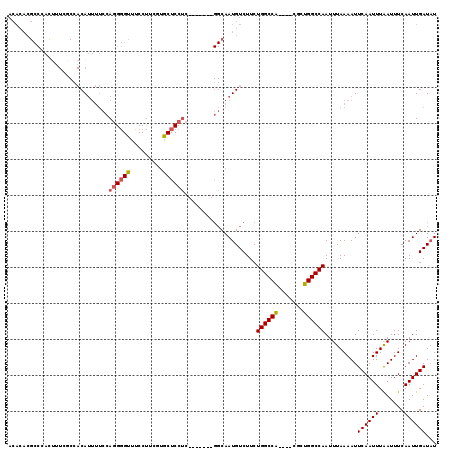
| Location | 18,800,980 – 18,801,087 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.99 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18800980 107 - 22407834 ACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUUAGCCCACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCUUCGUGCUCCUC-------GGCAAUGUCUUCUGGCCA ....((((((........((((...))))........))))))............((((((((.((.((((((.........)))))).-------)).))))).....))).. ( -29.29) >DroSec_CAF1 63488 107 - 1 ACAAGUGUGGCCAACUUCGAGCCACGCCCAUAUUAGCCCACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCUUCUUGCUCCUC-------GGCAAUGUCUUCUGGCCA ....((((((((......).)))))))........(((.......((........)).(((((.((.((((((.........)))))).-------)).))))).....))).. ( -26.00) >DroSim_CAF1 64115 107 - 1 ACAAGUGUGGCCAACUUCGAGCCACGCCCAUAUUAGCCCACACACGCCCACUUUGGCCACAUUUUCCAGGGGUUUCCUUCUUGCUCCUC-------GGCAAUGUCUUCUGGCCA ....((((((((......).)))))))..........................((((((((((.((.((((((.........)))))).-------)).))))).....))))) ( -29.70) >DroEre_CAF1 64276 107 - 1 ACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUCAGCCCACACACGCCCAUGUUUUCCACAUUUUCCAGGGGUUUCCUCCGUGCUCCUC-------GGCAAUGUCUUCUGGCCA ....((((((((.......))))))))........(((...(((.(((.((((.....)))).....((((((.........)))))).-------)))..))).....))).. ( -28.40) >DroYak_CAF1 64065 114 - 1 ACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUCAGCCCACACACGCCCAUUAUCCCGACAUUUUCCAGGGGUUUCCUUCGUACUCCUCGAAGUACGGCAAUGUCUUCUGGCCA ....((((((((.......))))))))........(((...(((.(((....(((((...........))))).......(((((......))))))))..))).....))).. ( -32.30) >consensus ACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUUAGCCCACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCUUCGUGCUCCUC_______GGCAAUGUCUUCUGGCCA ....((((((((.......))))))))........(((...(((.(((...................((((((.........))))))........)))..))).....))).. (-22.75 = -22.99 + 0.24)

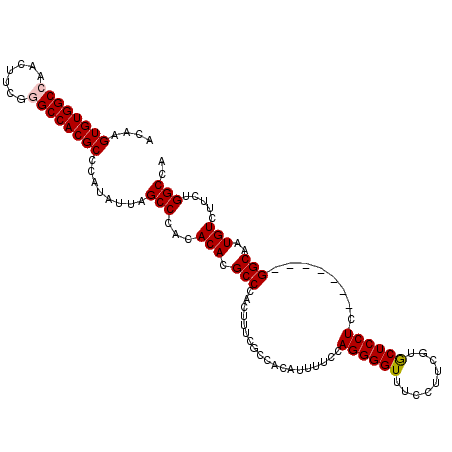
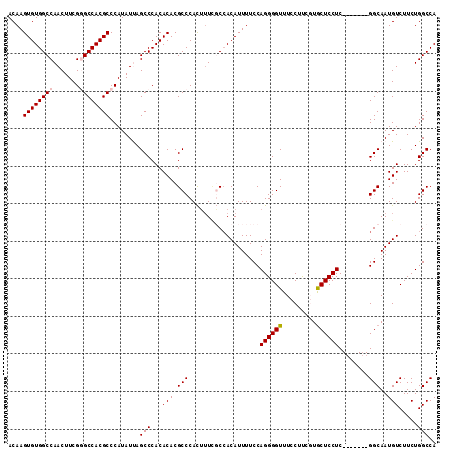
| Location | 18,801,009 – 18,801,115 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -33.46 |
| Energy contribution | -32.74 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18801009 106 + 22407834 AGGAAACCCCUGGAAAAUGUGGCGAAAGUGGGCGUGUGUGGGCUAAUAUGGGCGUGGCCCGAAGUUGGCCACACUUGUUUGUGACAGCCCAAGAGGGCAAUAAAGA .(....)((((..........((....))((((..((((((.(((((.(((((...)))))..))))))))))).((((...))))))))...))))......... ( -37.70) >DroSec_CAF1 63517 106 + 1 AGGAAACCCCUGGAAAAUGUGGCGAAAGUGGGCGUGUGUGGGCUAAUAUGGGCGUGGCUCGAAGUUGGCCACACUUGUUUGUGACAGCCCAAGAGGGCAAUAAAGC .(....)((((..........((....))((((..((((((.(((((.(((((...)))))..))))))))))).((((...))))))))...))))......... ( -34.70) >DroSim_CAF1 64144 106 + 1 AGGAAACCCCUGGAAAAUGUGGCCAAAGUGGGCGUGUGUGGGCUAAUAUGGGCGUGGCUCGAAGUUGGCCACACUUGUUUGUGACAGCCCAAGAGGGCAAUAAAGC (((.....)))......(((.(((....(((((..((((((.(((((.(((((...)))))..))))))))))).((((...)))))))))....))).))).... ( -35.50) >DroEre_CAF1 64305 106 + 1 AGGAAACCCCUGGAAAAUGUGGAAAACAUGGGCGUGUGUGGGCUGAUAUGGGCGUGGCCCGAAGUUGGCCACACUUGUUUGUGACAGCCCAAGAGGGCAAUAAAGC (((.....)))...............(((..(((.((((((.(..((.(((((...)))))..))..))))))).)))..)))...((((....))))........ ( -37.00) >DroYak_CAF1 64101 106 + 1 AGGAAACCCCUGGAAAAUGUCGGGAUAAUGGGCGUGUGUGGGCUGAUAUGGGCGUGGCCCGAAGUUGGCCACACUUGUUUGUGACAGCCCAAGAGGGCAAUAAAGC .(....).(((((......)))))...((..(((.((((((.(..((.(((((...)))))..))..))))))).)))..))....((((....))))........ ( -37.70) >consensus AGGAAACCCCUGGAAAAUGUGGCGAAAGUGGGCGUGUGUGGGCUAAUAUGGGCGUGGCCCGAAGUUGGCCACACUUGUUUGUGACAGCCCAAGAGGGCAAUAAAGC (((.....)))................(..((((.((((((.(((((.(((((...)))))..))))))))))).))))..)....((((....))))........ (-33.46 = -32.74 + -0.72)


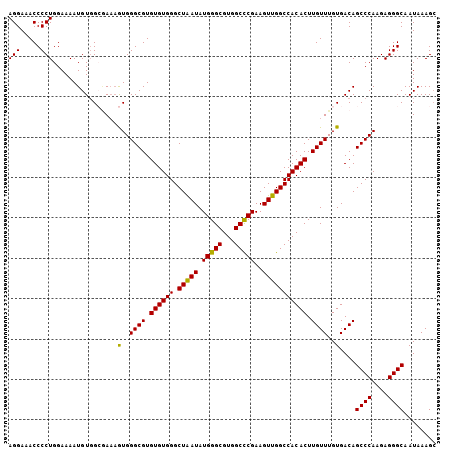
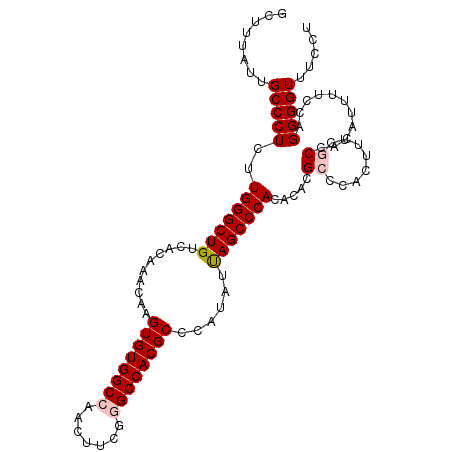
| Location | 18,801,009 – 18,801,115 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.89 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18801009 106 - 22407834 UCUUUAUUGCCCUCUUGGGCUGUCACAAACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUUAGCCCACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCU ........(((((..(((((((..........((((((((.......))))))))......))))))).....((........))...........)))))..... ( -30.19) >DroSec_CAF1 63517 106 - 1 GCUUUAUUGCCCUCUUGGGCUGUCACAAACAAGUGUGGCCAACUUCGAGCCACGCCCAUAUUAGCCCACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCU ........(((((..(((((((..........((((((((......).)))))))......))))))).....((........))...........)))))..... ( -26.99) >DroSim_CAF1 64144 106 - 1 GCUUUAUUGCCCUCUUGGGCUGUCACAAACAAGUGUGGCCAACUUCGAGCCACGCCCAUAUUAGCCCACACACGCCCACUUUGGCCACAUUUUCCAGGGGUUUCCU ........(((((..(((((((..........((((((((......).)))))))......))))))).....(((......)))...........)))))..... ( -30.39) >DroEre_CAF1 64305 106 - 1 GCUUUAUUGCCCUCUUGGGCUGUCACAAACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUCAGCCCACACACGCCCAUGUUUUCCACAUUUUCCAGGGGUUUCCU ........(((((..(((((((..........((((((((.......))))))))......))))))).........((((.....))))......)))))..... ( -31.99) >DroYak_CAF1 64101 106 - 1 GCUUUAUUGCCCUCUUGGGCUGUCACAAACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUCAGCCCACACACGCCCAUUAUCCCGACAUUUUCCAGGGGUUUCCU ((......)).....(((((((..........((((((((.......))))))))......)))))))............(((((...........)))))..... ( -31.59) >consensus GCUUUAUUGCCCUCUUGGGCUGUCACAAACAAGUGUGGCCAACUUCGGGCCACGCCCAUAUUAGCCCACACACGCCCACUUUCGCCACAUUUUCCAGGGGUUUCCU ........(((((..(((((((..........((((((((.......))))))))......))))))).....((........))...........)))))..... (-27.33 = -27.89 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:35 2006