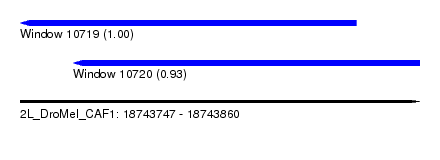
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,743,747 – 18,743,860 |
| Length | 113 |
| Max. P | 0.999014 |

| Location | 18,743,747 – 18,743,842 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18743747 95 - 22407834 CCAGUGAGCAGUGAGCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGCUUAAUUACGAACAAG ((((((((((.(....).))))).(((((((.........))))))).....))))).(((((((((....)))))))))............... ( -33.10) >DroSec_CAF1 7414 88 - 1 CCAGUGA-------GCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGCUUAAUUACGAACAAG (((((((-------((...)))).(((((((.........))))))).....))))).(((((((((....)))))))))............... ( -29.30) >DroSim_CAF1 7590 88 - 1 CCCGUGA-------GCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACCGGCGCAUGUGCCACACGGCGUAUGCUUAAUUACGAACAAG ..(((((-------((..((....(((((((.........)))))))...))....))(((((((((....)))))))))....)))))...... ( -28.10) >DroEre_CAF1 7615 88 - 1 CCAGUGA-------GCAGUGUUCAGCUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGCUUAAUUACGAACAAG (((((((-------(((((.....)))))...........((....)).)).))))).(((((((((....)))))))))............... ( -26.70) >DroYak_CAF1 7504 88 - 1 CCAUUGA-------GCAGUGUUCAGCUGCCAACUAGAAACUGGCUACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGCUUAAUUACGAACAAG (((((((-------(((((.....)))))...((((...))))......)))).))).(((((((((....)))))))))............... ( -23.00) >consensus CCAGUGA_______GCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGCUUAAUUACGAACAAG ..................(((((.(((((((.........)))))))...........(((((((((....)))))))))........))))).. (-25.04 = -25.64 + 0.60)

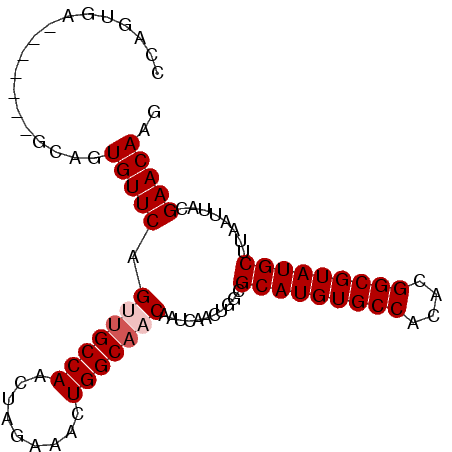
| Location | 18,743,762 – 18,743,860 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.62 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18743762 98 - 22407834 UUGUUGGCAUCCCAGUGCCCAGUGAGCAGUGAGCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGC .(((((((((....((((((((((((((.(....).))))).(((((((.........))))))).....))))).)))))))))).)))........ ( -36.20) >DroSec_CAF1 7429 91 - 1 UUGUUGGCAUCCCAGUGCCCAGUGA-------GCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGC .(((((((((....(((((((((((-------((...)))).(((((((.........))))))).....))))).)))))))))).)))........ ( -32.40) >DroSim_CAF1 7605 91 - 1 UUGUUGGCAUCCCAGUGCCCCGUGA-------GCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACCGGCGCAUGUGCCACACGGCGUAUGC .....(((((....)))))((((((-------((...)))))(((((((.........))))))).......))).(((((((((....))))))))) ( -31.30) >DroEre_CAF1 7630 91 - 1 UGGCUGGCAUCCCAGUGCCCAGUGA-------GCAGUGUUCAGCUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGC ..(((((....)))))(((.(((((-------(((((.....)))))...........((....)).)).))))))(((((((((....))))))))) ( -32.10) >DroYak_CAF1 7519 91 - 1 UUGUUGGCAUCCCAGUGCCCAUUGA-------GCAGUGUUCAGCUGCCAACUAGAAACUGGCUACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGC ..(((((....(((((.....(((.-------(((((.....))))))))......)))))......)))))....(((((((((....))))))))) ( -28.40) >consensus UUGUUGGCAUCCCAGUGCCCAGUGA_______GCAGUGUUCAGUUGCCAACUAGAAACUGGCAACAAUCAACUGGCGCAUGUGCCACACGGCGUAUGC ..((.(((((....)))))..............((((.....(((((((.........))))))).....))))))(((((((((....))))))))) (-26.98 = -27.62 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:12 2006