
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,970,630 – 1,970,730 |
| Length | 100 |
| Max. P | 0.987513 |

| Location | 1,970,630 – 1,970,730 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -39.72 |
| Consensus MFE | -23.38 |
| Energy contribution | -25.46 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
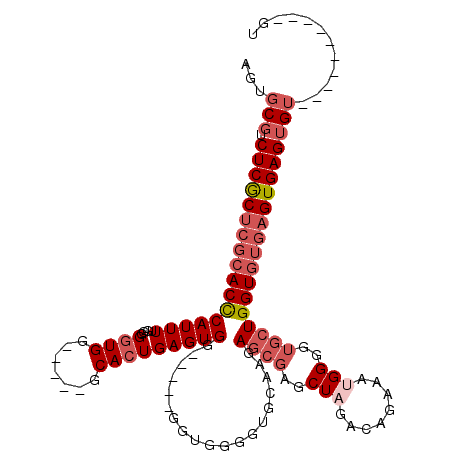
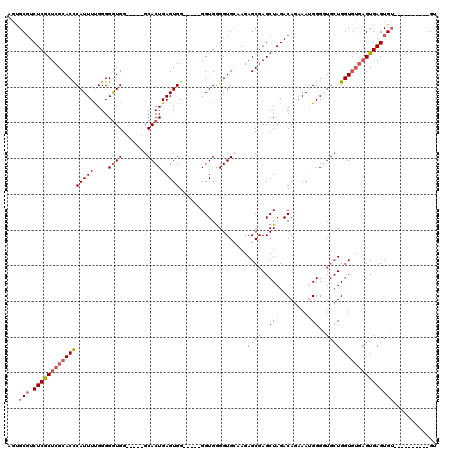
>2L_DroMel_CAF1 1970630 100 + 22407834 AAUGCGUCUCACUCGCACUCAUUUUGGGGGUGG-----GCACUGAGUGG-----GGUGGGGUGCAAGAGCGAGCUAGACAGAAAUGGGGUGCUGGUGUGAGUGAGUGU----------GU .(((((.((((((((((((((((.....))))(-----(((((..((.(-----..(((..(((....)))..)))..)....))..)))))))))))))))))))))----------)) ( -37.60) >DroSec_CAF1 13077 100 + 1 AGUGCGUCUCGCUCGCACCCAUUUUGGGGGUGG-----GCACUGAGUGG-----GGUGGGGUGCAAGAGCGAGCUAGACAGAAAUGGGGUGCUGGUGUGAGUGAGUGU----------GU .(..((.((((((((((((((((.....))))(-----(((((..((.(-----..(((..(((....)))..)))..)....))..)))))))))))))))))))).----------.) ( -39.50) >DroSim_CAF1 17123 100 + 1 AGUGCGUCUCGCUCGCACCCAUUUUGGGGGUGG-----GCACUGAGUGG-----GGUGGGGUGCAAGAGCGAGCUAGACAGAAAUGGGGUGCUGGUGUGAGUGAGUGU----------GU .(..((.((((((((((((((((.....))))(-----(((((..((.(-----..(((..(((....)))..)))..)....))..)))))))))))))))))))).----------.) ( -39.50) >DroEre_CAF1 22243 112 + 1 AACGCGUCUCGCUCGCACCCAUUUCGGGGAUGGGAUAAGCACUGAGUGG-----GGUGGGGUGAAAGAGCGAGCUAGACAAAAAGGGGC---UGGUGUGAGUGAGUGUGUGUGUGUGUGC .(((((.(((((((((.(((((..((....(((........)))..)).-----.)))))........((.((((...(.....).)))---).)))))))))))))))).......... ( -37.90) >DroYak_CAF1 20254 109 + 1 AGCGCCUCUCGCUCGCACCCAUUUCGGGGGUGC-----GCACUGAGUGGAGUGGGGUGAGGUGAAAGAGCGAGCUAGACAGCAAGGGGCUGCUGGU----GUGAGUG--UGUGUGUGUGU .(((((((((((((((((((.......))))))-----.((((......)))).............))))))(((((.((((.....)))))))))----..))).)--)))........ ( -44.10) >consensus AGUGCGUCUCGCUCGCACCCAUUUUGGGGGUGG_____GCACUGAGUGG_____GGUGGGGUGCAAGAGCGAGCUAGACAGAAAUGGGGUGCUGGUGUGAGUGAGUGU__________GU ...(((.(((((((((((((((((....((((.......)))))))))...................((((..(((........)))..)))))))))))))))))))............ (-23.38 = -25.46 + 2.08)

| Location | 1,970,630 – 1,970,730 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
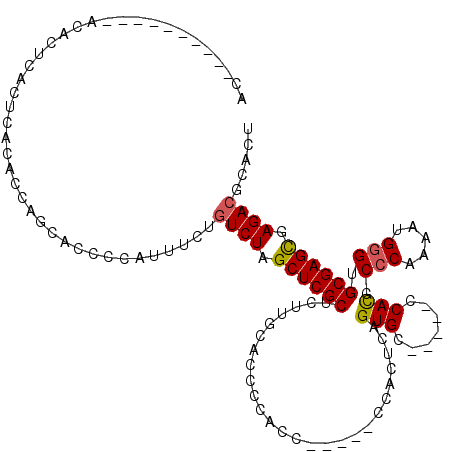
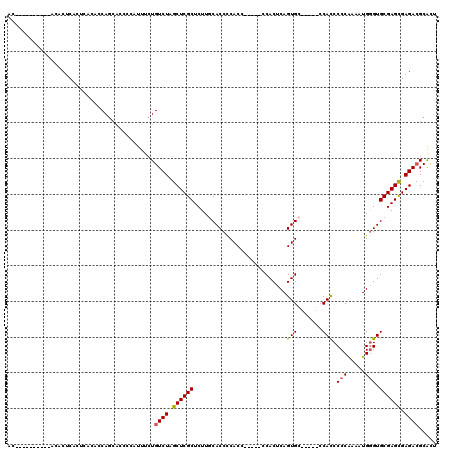
>2L_DroMel_CAF1 1970630 100 - 22407834 AC----------ACACUCACUCACACCAGCACCCCAUUUCUGUCUAGCUCGCUCUUGCACCCCACC-----CCACUCAGUGC-----CCACCCCCAAAAUGAGUGCGAGUGAGACGCAUU ..----------...(((((((............................((....))........-----.((((((.((.-----.......))...)))))).)))))))....... ( -21.00) >DroSec_CAF1 13077 100 - 1 AC----------ACACUCACUCACACCAGCACCCCAUUUCUGUCUAGCUCGCUCUUGCACCCCACC-----CCACUCAGUGC-----CCACCCCCAAAAUGGGUGCGAGCGAGACGCACU ..----------.............................((((.((((((....((((......-----.......))))-----..((((.......)))))))))).))))..... ( -24.52) >DroSim_CAF1 17123 100 - 1 AC----------ACACUCACUCACACCAGCACCCCAUUUCUGUCUAGCUCGCUCUUGCACCCCACC-----CCACUCAGUGC-----CCACCCCCAAAAUGGGUGCGAGCGAGACGCACU ..----------.............................((((.((((((....((((......-----.......))))-----..((((.......)))))))))).))))..... ( -24.52) >DroEre_CAF1 22243 112 - 1 GCACACACACACACACUCACUCACACCA---GCCCCUUUUUGUCUAGCUCGCUCUUUCACCCCACC-----CCACUCAGUGCUUAUCCCAUCCCCGAAAUGGGUGCGAGCGAGACGCGUU ((.................((......)---).........((((.((((((..........(((.-----.......))).....(((((.......))))).)))))).))))))... ( -23.00) >DroYak_CAF1 20254 109 - 1 ACACACACACA--CACUCAC----ACCAGCAGCCCCUUGCUGUCUAGCUCGCUCUUUCACCUCACCCCACUCCACUCAGUGC-----GCACCCCCGAAAUGGGUGCGAGCGAGAGGCGCU ...........--......(----.((.(((((.....)))))....(((((((.............((((......)))).-----((((((.......))))))))))))).)).).. ( -33.10) >consensus AC__________ACACUCACUCACACCAGCACCCCAUUUCUGUCUAGCUCGCUCUUGCACCCCACC_____CCACUCAGUGC_____CCACCCCCAAAAUGGGUGCGAGCGAGACGCACU .........................................((((.((((((..........................(((.......))).(((.....))).)))))).))))..... (-18.50 = -18.58 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:32 2006