
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,733,630 – 18,733,870 |
| Length | 240 |
| Max. P | 0.998154 |

| Location | 18,733,630 – 18,733,750 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18733630 120 - 22407834 AUAAUUAAAUAUGUAAGAGGCAAGAGCCCAAGUUAAUAACAGCUGCUACCACAGAAGCCUUUUGCCGGCCAAACGUUAAAAAUUUAAUAAUCGCCUGCGAGUAUAAGGCGGCCAGAUGCA ...........((((...((((((((....((((......))))(((........)))))))))))((((....(((((....)))))....((((.........))))))))...)))) ( -29.80) >DroSec_CAF1 26155 120 - 1 AUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAACAGCUGCUACCACAGAAGCCUUUUGCCGGCCAAACGCUGAAAAUUUAAUAAUCGCCUGCGAGUAUAAGGCGGCCAGAUGCA ...(((((((..(((((((((.(((......)))........(((......)))..)))))))))((((.....))))...)))))))..((((((.........))))))......... ( -29.00) >DroSim_CAF1 24710 120 - 1 AUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAACAGCUGCUACCACAGAAGCCUUUUGCCGGCCAAACGCUGAAAAUUUAAUAAUCGCCUGCGAGUACAAGGCGGCCAGAUGCA ...(((((((..(((((((((.(((......)))........(((......)))..)))))))))((((.....))))...)))))))..((((((.........))))))......... ( -29.00) >DroEre_CAF1 26184 120 - 1 AUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAACAGCUGCUACCACAGAAGCCUUUUGCCGGCCAAACGCUGAAAAUUUAAUAAUCGCCUGCGAGUACAAAGCGGCCAGUUGCA ...(((((((..(((((((((.(((......)))........(((......)))..)))))))))((((.....))))...)))))))....(((.((.........)))))........ ( -26.60) >DroYak_CAF1 26257 120 - 1 AUAAUUAAAUAUGUAAGAGGCAAGCGACCAAGUUAAUAACAGCUGCUACCACAGAAGCCUUUUGCCGGCCAAACGCUGAAAAUUUAAUAAUCGCCUGCGAGCAUAAGGCGGCCAGAUGCA ...(((((((..(((((((((.(((......)))........(((......)))..)))))))))((((.....))))...)))))))..((((((.........))))))......... ( -29.00) >consensus AUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAACAGCUGCUACCACAGAAGCCUUUUGCCGGCCAAACGCUGAAAAUUUAAUAAUCGCCUGCGAGUAUAAGGCGGCCAGAUGCA ...(((((((..(((((((((.(((......)))........(((......)))..)))))))))((((.....))))...)))))))..((((((.........))))))......... (-25.38 = -25.82 + 0.44)

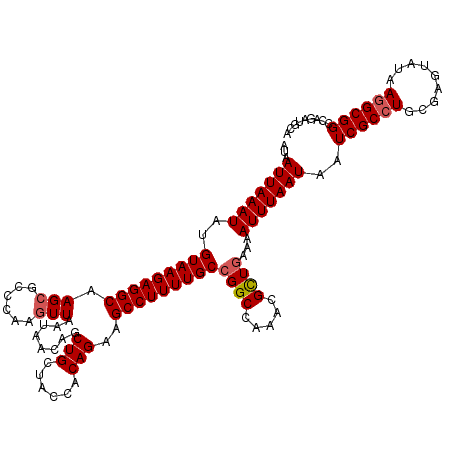

| Location | 18,733,710 – 18,733,830 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -42.76 |
| Consensus MFE | -38.58 |
| Energy contribution | -38.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18733710 120 + 22407834 GUUAUUAACUUGGGCUCUUGCCUCUUACAUAUUUAAUUAUUUGGGCGGGCCAGGUCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGG .........((((((.............(((......))).(((((.((((..((((..(....)..))))((((((((...)))))))).........))))...)))))))))))... ( -42.40) >DroSec_CAF1 26235 120 + 1 GUUAUUAACUUGGGCGCUUGCCUCUUACAUAUUUAAUUAUUUGGGCGGGCCAGGUCUUUAAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGA .........((((((((((((((.......((((((..((((((.....))))))..))))))....)))))))).(((.....(((((.((....)).)))))...))).))))))... ( -43.50) >DroSim_CAF1 24790 120 + 1 GUUAUUAACUUGGGCGCUUGCCUCUUACAUAUUUAAUUAUUUGGGCGGGCCAGGUCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGA .........((((((((((((((.......((((((..((((((.....))))))..))))))....)))))))).(((.....(((((.((....)).)))))...))).))))))... ( -43.60) >DroEre_CAF1 26264 118 + 1 GUUAUUAACUUGGGCGCUUGCCUCUUACAUAUUUAAUUAUUUGG--GGGCCAGGCCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUUGGCCACAGCCCAGCCCAAUGA .........((((((((((((((((...(((......)))..))--))).)))))...........(((((((((((((...))))))))...((.....))....)))))))))))... ( -44.70) >DroYak_CAF1 26337 118 + 1 GUUAUUAACUUGGUCGCUUGCCUCUUACAUAUUUAAUUAUUUGG--GGGCCAGGCCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUUGGCCACAGCCCAGCCCAAUGA ................((.((((((...(((......)))..))--)))).))((((..(....)..))))((((.(((.....(((((..........)))))...))).))))..... ( -39.60) >consensus GUUAUUAACUUGGGCGCUUGCCUCUUACAUAUUUAAUUAUUUGGGCGGGCCAGGUCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGA .........((((((((((((((((((.(((......))).)))).))).)))))...........(((((.((((((((((.(......).)).))))))))...)))))))))))... (-38.58 = -38.98 + 0.40)

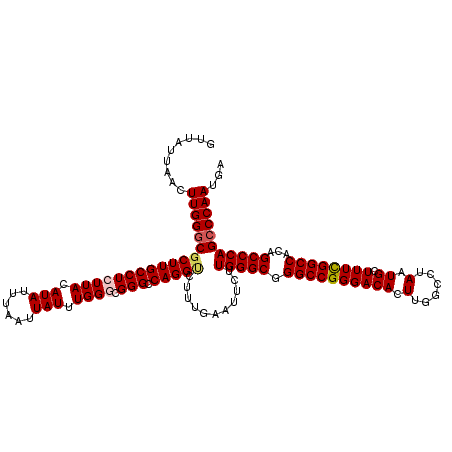
| Location | 18,733,710 – 18,733,830 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.82 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18733710 120 - 22407834 CCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGACCUGGCCCGCCCAAAUAAUUAAAUAUGUAAGAGGCAAGAGCCCAAGUUAAUAAC ...(((((((((((.(.(((((...(((((......)))))..))))))))))..............((.((((...((.((......)).))...).))).)))))))))......... ( -36.90) >DroSec_CAF1 26235 120 - 1 UCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUUAAAGACCUGGCCCGCCCAAAUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAAC ...(((((((((((.(.(((((...(((((......)))))..)))))))))))................((..((((..(((........)))..).)))..))))))))......... ( -37.10) >DroSim_CAF1 24790 120 - 1 UCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGACCUGGCCCGCCCAAAUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAAC ...(((((((((((.(.(((((...(((((......)))))..)))))))))))................((..((((..(((........)))..).)))..))))))))......... ( -37.10) >DroEre_CAF1 26264 118 - 1 UCAUUGGGCUGGGCUGUGGCCAAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGGCCUGGCCC--CCAAAUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAAC ((.((((((.(((((((((((..........))))).......)))))))))))))).......(((((.((((--....(((......)))....).))).)).)))............ ( -37.91) >DroYak_CAF1 26337 118 - 1 UCAUUGGGCUGGGCUGUGGCCAAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGGCCUGGCCC--CCAAAUAAUUAAAUAUGUAAGAGGCAAGCGACCAAGUUAAUAAC ((.((((((.(((((((((((..........))))).......)))))))))))))).......(((((.((((--....(((......)))....).))).)).).))........... ( -35.01) >consensus UCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGACCUGGCCCGCCCAAAUAAUUAAAUAUGUAAGAGGCAAGCGCCCAAGUUAAUAAC ...(((((((((((.(.(((((...(((((......)))))..))))))))))).............((.((((...((.((......)).))...).))).)).))))))......... (-32.22 = -32.82 + 0.60)

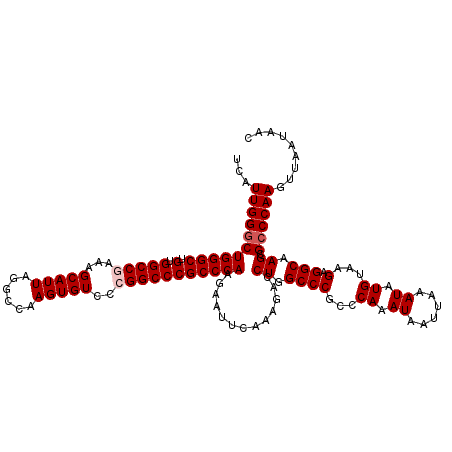

| Location | 18,733,750 – 18,733,870 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -43.14 |
| Consensus MFE | -39.46 |
| Energy contribution | -39.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18733750 120 + 22407834 UUGGGCGGGCCAGGUCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGGCCAACCUAUUGGCCUUAAUAUCUGCUGUCGAUCUGUCCAU .(((((((((((((..........)))))((((((.(((.....(((((.((....)).)))))...))).))))...((((((....)))))).........))......)))))))). ( -48.10) >DroSec_CAF1 26275 120 + 1 UUGGGCGGGCCAGGUCUUUAAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGACCAACCUAUUGGCCUUAAUAUCCGCUGUCGAUCUGUCCAU .(((((((..(.((((.........((((((((((.((....))(((((.((....)).)))))..)))).)))))).))))..((....)).................)..))))))). ( -41.50) >DroSim_CAF1 24830 120 + 1 UUGGGCGGGCCAGGUCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGACCAACCUAUUGGCCUUAAUAUCCGCUGUCGAUCUGUCCAU .((((((((((((((((..(....)..))))((((.(((.....(((((.((....)).)))))...))).)))).............))))))).......((....))....))))). ( -42.90) >DroEre_CAF1 26304 118 + 1 UUGG--GGGCCAGGCCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUUGGCCACAGCCCAGCCCAAUGACCAACCUAUUGGCCUUAAUAUCCGCUGUCGAUUCGUCCAU .(((--(((((((((((..(....)..))))((((.(((.....(((((..........)))))...))).)))).............))))))).......((....))......))). ( -41.60) >DroYak_CAF1 26377 118 + 1 UUGG--GGGCCAGGCCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUUGGCCACAGCCCAGCCCAAUGACCAACCUAUUGGCCUUAAUAUCCGCUGUCGAUUUGUCCAU .(((--(((((((((((..(....)..))))((((.(((.....(((((..........)))))...))).)))).............))))))).......((....))......))). ( -41.60) >consensus UUGGGCGGGCCAGGUCUUUGAAUUCUUGGGCGGGCCGGGACACUUGGCCUAAUGCUUUCGGCCACAGCCCAGCCCAAUGACCAACCUAUUGGCCUUAAUAUCCGCUGUCGAUCUGUCCAU .((((((((((((((((..(....)..))))((((.(((.....(((((.((....)).)))))...))).)))).............))))))........((....))...)))))). (-39.46 = -39.90 + 0.44)


| Location | 18,733,750 – 18,733,870 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -43.93 |
| Consensus MFE | -42.67 |
| Energy contribution | -42.27 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18733750 120 - 22407834 AUGGACAGAUCGACAGCAGAUAUUAAGGCCAAUAGGUUGGCCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGACCUGGCCCGCCCAA ..((.(((.((...............((((((....)))))).((((((.(((((((((((..........))))).......)))))))))))).........)).)))..))...... ( -45.51) >DroSec_CAF1 26275 120 - 1 AUGGACAGAUCGACAGCGGAUAUUAAGGCCAAUAGGUUGGUCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUUAAAGACCUGGCCCGCCCAA .(((.(...(((....))).......(((((...((((..((.((((((.(((((((((((..........))))).......)))))))))))))).......))))))))).).))). ( -43.81) >DroSim_CAF1 24830 120 - 1 AUGGACAGAUCGACAGCGGAUAUUAAGGCCAAUAGGUUGGUCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGACCUGGCCCGCCCAA .(((.(...(((....))).......(((((...((((.(...((((((.(((((((((((..........))))).......)))))))))))).....)...))))))))).).))). ( -43.81) >DroEre_CAF1 26304 118 - 1 AUGGACGAAUCGACAGCGGAUAUUAAGGCCAAUAGGUUGGUCAUUGGGCUGGGCUGUGGCCAAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGGCCUGGCCC--CCAA .(((..(((((....((.........((((((....))))))...(((((((((..(((((..........)))))...).))))))))))....).))))...(((...))).--))). ( -43.90) >DroYak_CAF1 26377 118 - 1 AUGGACAAAUCGACAGCGGAUAUUAAGGCCAAUAGGUUGGUCAUUGGGCUGGGCUGUGGCCAAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGGCCUGGCCC--CCAA .(((.....(((....))).......(((((.....((((...((((((.(((((((((((..........))))).......))))))))))))....)))).....))))).--))). ( -42.61) >consensus AUGGACAGAUCGACAGCGGAUAUUAAGGCCAAUAGGUUGGUCAUUGGGCUGGGCUGUGGCCGAAAGCAUUAGGCCAAGUGUCCCGGCCCGCCCAAGAAUUCAAAGACCUGGCCCGCCCAA .(((....(((.......))).....(((((...((((.(...((((((.(((((((((((..........))))).......)))))))))))).....)...)))))))))...))). (-42.67 = -42.27 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:05 2006