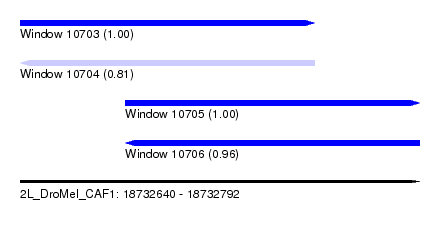
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,732,640 – 18,732,792 |
| Length | 152 |
| Max. P | 0.998231 |

| Location | 18,732,640 – 18,732,752 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -44.54 |
| Consensus MFE | -35.96 |
| Energy contribution | -38.56 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18732640 112 + 22407834 CCAUCGUUGUCGGUUCCACAUUGCACCACUGGGUUUUUAAAGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGAGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAA .....(((((((.((((((((((((((.((..(((.....)))..)).).))))((((((((((....))))))))..((((.....))))--------))))))))))))))))))... ( -42.80) >DroSec_CAF1 25189 112 + 1 CCAUGGUUGUCGGUUCCACAUUGCACCACUGGGUUUUAAAAGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAA .....(((((((.((((((((((((((.((..(((.....)))..)).).))))((((((((((....))))))))))((((.....))))--------..))))))))))))))))... ( -47.10) >DroSim_CAF1 23731 112 + 1 CCAUGGUUGUCGGUUCCACAUUGCACCACUGGGUUUUAAAAGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAA .....(((((((.((((((((((((((.((..(((.....)))..)).).))))((((((((((....))))))))))((((.....))))--------..))))))))))))))))... ( -47.10) >DroEre_CAF1 25286 108 + 1 CCAU----GGCGGUUCCACUGUGCACCACUGGGUUGUAAAAGCGAAGAGUGUGCCCCAGCAGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAA ....----.((.(((((((...((((.(((...((((....))))..)))))))((((((.(((....))).))))))((((.....))))--------.....))))))).))...... ( -38.50) >DroYak_CAF1 25254 120 + 1 CCAUCGUUGUCGGUUCCACUGUGCUCCAGUGGGUUUUAAAAGCGAAGUGUGUGCCCCAGCAGAAAAGAUUCAGUUGGGUGGCUAUCUGCCACCCAAUGUGGAAUGUGGAAUGGCGGCAAA ...(((((......(((((((.....))))))).......)))))......(((((((.......(.((((.((((((((((.....))))))))))...)))).)....))).)))).. ( -47.22) >consensus CCAUCGUUGUCGGUUCCACAUUGCACCACUGGGUUUUAAAAGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA________GGAAUGUGGAAUGGCGGCAAA .....(((((((.((((((((((((((.((..(((.....)))..)).).))))((((((((((....))))))))))((((.....))))..........))))))))))))))))... (-35.96 = -38.56 + 2.60)

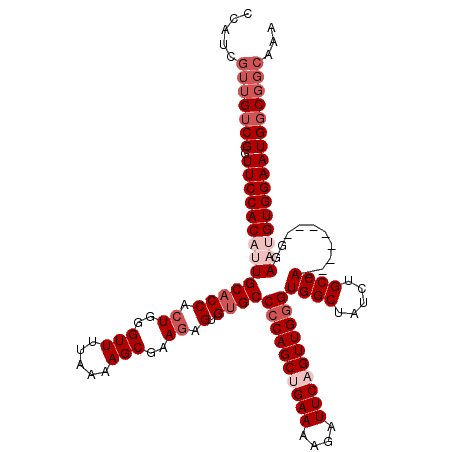

| Location | 18,732,640 – 18,732,752 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -32.79 |
| Consensus MFE | -25.65 |
| Energy contribution | -26.89 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18732640 112 - 22407834 UUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACUCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCUUUAAAAACCCAGUGGUGCAAUGUGGAACCGACAACGAUGG .(((.((...(((((((((..--------((((.....))))((((.(((((....))))).))))(((((((.................))).))))))))))))).)).)))...... ( -30.93) >DroSec_CAF1 25189 112 - 1 UUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACCCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCUUUUAAAACCCAGUGGUGCAAUGUGGAACCGACAACCAUGG .(((.((...(((((((((..--------((((.....))))((((.(((((....))))).))))(((((((.................))).))))))))))))).)).)))...... ( -33.93) >DroSim_CAF1 23731 112 - 1 UUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACCCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCUUUUAAAACCCAGUGGUGCAAUGUGGAACCGACAACCAUGG .(((.((...(((((((((..--------((((.....))))((((.(((((....))))).))))(((((((.................))).))))))))))))).)).)))...... ( -33.93) >DroEre_CAF1 25286 108 - 1 UUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACCCAACUGAAUCUUUUCUGCUGGGGCACACUCUUCGCUUUUACAACCCAGUGGUGCACAGUGGAACCGCC----AUGG ......((..((((((...((--------..(((((.....................)))))..))(((((((.................))).))))...))))))..)).----.... ( -28.43) >DroYak_CAF1 25254 120 - 1 UUUGCCGCCAUUCCACAUUCCACAUUGGGUGGCAGAUAGCCACCCAACUGAAUCUUUUCUGCUGGGGCACACACUUCGCUUUUAAAACCCACUGGAGCACAGUGGAACCGACAACGAUGG ..((((.(((..............(((((((((.....)))))))))..(((....)))...)))))))....(.(((..........((((((.....)))))).........))).). ( -36.71) >consensus UUUGCCGCCAUUCCACAUUCC________UGGCAGAUAGCCACCCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCUUUUAAAACCCAGUGGUGCAAUGUGGAACCGACAACCAUGG .(((.((...((((((.............((((.....))))((((.(((((....))))).))))(((((((.................))).))))...)))))).)).)))...... (-25.65 = -26.89 + 1.24)

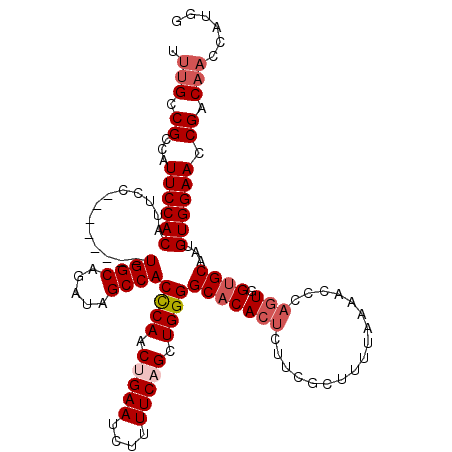
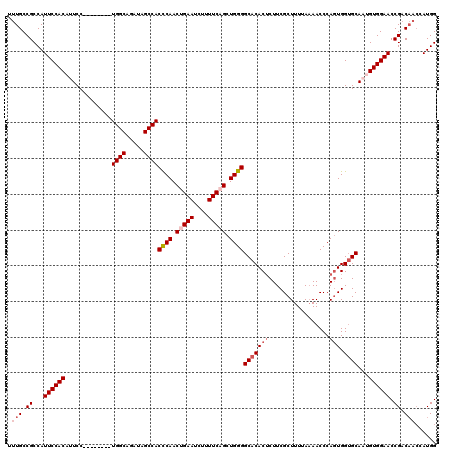
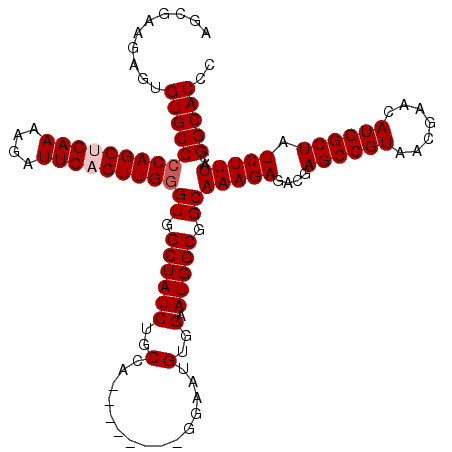
| Location | 18,732,680 – 18,732,792 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -40.54 |
| Consensus MFE | -36.84 |
| Energy contribution | -37.44 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18732680 112 + 22407834 AGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGAGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAAGAGACGAGCCGUAACGAACAUGGCUAUCUUUACGGCACCC ..........(((((.((((((((....)))))))).((.((((((..(..--------.....)..).))))).))(((((....((((((.......)))))).)))))..))))).. ( -37.90) >DroSec_CAF1 25229 112 + 1 AGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAAGAGACGAGCCGUAACGAACAUGGCUAUCUUUACGGCACCC ..........((((((((((((((....)))))))))((.((((((..(..--------.....)..).))))).))(((((....((((((.......)))))).)))))..))))).. ( -41.20) >DroSim_CAF1 23771 112 + 1 AGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAAGAGACGAGCCGUAACGAACAUGGCUAUCUUUACGGCACCC ..........((((((((((((((....)))))))))((.((((((..(..--------.....)..).))))).))(((((....((((((.......)))))).)))))..))))).. ( -41.20) >DroEre_CAF1 25322 112 + 1 AGCGAAGAGUGUGCCCCAGCAGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA--------GGAAUGUGGAAUGGCGGCAAAGAGACGAGCCGUAACGAACAUGGCUAUCUUUACGGCACCC ..........((((((((((.(((....))).)))))((.((((((..(..--------.....)..).))))).))(((((....((((((.......)))))).)))))..))))).. ( -35.90) >DroYak_CAF1 25294 120 + 1 AGCGAAGUGUGUGCCCCAGCAGAAAAGAUUCAGUUGGGUGGCUAUCUGCCACCCAAUGUGGAAUGUGGAAUGGCGGCAAAGAGACGAGCCGUAACGAACAUGGCUAUCUUUACGGCACCC ......((((.(((((((.......(.((((.((((((((((.....))))))))))...)))).)....))).)))).(((((..((((((.......)))))).)))))...)))).. ( -46.50) >consensus AGCGAAGAGUGUGCCCCAGCUGAAAAGAUUCAGUUGGGUGGCUAUCUGCCA________GGAAUGUGGAAUGGCGGCAAAGAGACGAGCCGUAACGAACAUGGCUAUCUUUACGGCACCC ..........((((((((((((((....)))))))))((.((((((..(...............)..).))))).))(((((....((((((.......)))))).)))))..))))).. (-36.84 = -37.44 + 0.60)


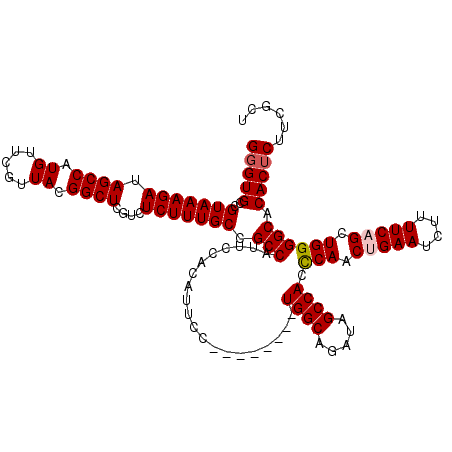
| Location | 18,732,680 – 18,732,792 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -31.80 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18732680 112 - 22407834 GGGUGCCGUAAAGAUAGCCAUGUUCGUUACGGCUCGUCUCUUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACUCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCU (((((..(((((((.((((.((.....)).))))....))))))).(((............--------((((.....)))).(((.(((((....))))).)))))).)))))...... ( -32.30) >DroSec_CAF1 25229 112 - 1 GGGUGCCGUAAAGAUAGCCAUGUUCGUUACGGCUCGUCUCUUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACCCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCU (((((..(((((((.((((.((.....)).))))....))))))).(((............--------((((.....)))).(((.(((((....))))).)))))).)))))...... ( -35.30) >DroSim_CAF1 23771 112 - 1 GGGUGCCGUAAAGAUAGCCAUGUUCGUUACGGCUCGUCUCUUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACCCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCU (((((..(((((((.((((.((.....)).))))....))))))).(((............--------((((.....)))).(((.(((((....))))).)))))).)))))...... ( -35.30) >DroEre_CAF1 25322 112 - 1 GGGUGCCGUAAAGAUAGCCAUGUUCGUUACGGCUCGUCUCUUUGCCGCCAUUCCACAUUCC--------UGGCAGAUAGCCACCCAACUGAAUCUUUUCUGCUGGGGCACACUCUUCGCU (((((..(((((((.((((.((.....)).))))....))))))).(((............--------((((.....)))).(((.(.(((....))).).)))))).)))))...... ( -30.70) >DroYak_CAF1 25294 120 - 1 GGGUGCCGUAAAGAUAGCCAUGUUCGUUACGGCUCGUCUCUUUGCCGCCAUUCCACAUUCCACAUUGGGUGGCAGAUAGCCACCCAACUGAAUCUUUUCUGCUGGGGCACACACUUCGCU ..((((((((((((.((((.((.....)).))))....)))))))..............((((((((((((((.....)))))))))..(((....))))).)))))))).......... ( -39.90) >consensus GGGUGCCGUAAAGAUAGCCAUGUUCGUUACGGCUCGUCUCUUUGCCGCCAUUCCACAUUCC________UGGCAGAUAGCCACCCAACUGAAUCUUUUCAGCUGGGGCACACUCUUCGCU (((((..(((((((.((((.((.....)).))))....))))))).(((....................((((.....)))).(((.(((((....))))).)))))).)))))...... (-31.80 = -32.24 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:59 2006