
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,731,217 – 18,731,377 |
| Length | 160 |
| Max. P | 0.979587 |

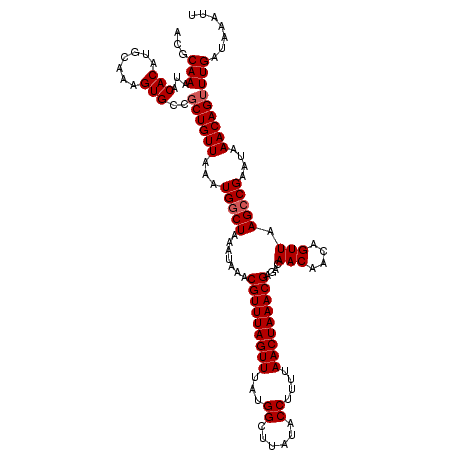
| Location | 18,731,217 – 18,731,337 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18731217 120 + 22407834 ACGCAAAUACACAAGCAAAGUGCCCCUGUUAAAUGGCUAAAUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACGACAGUUUAGCCGAAUAAACAGUUUGAUAAAUU ...((((..(((.......)))...(((((...(((((((((...(((((((((...((......))....))))))))).(....)....)))))))))....)))))))))....... ( -25.30) >DroSec_CAF1 23845 120 + 1 ACUCAAAUACACAUGCAAAGUGCCGCUGUUAAAUGGCUAAAUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUU ..((((...(((.......)))..((((((...(((((.......(((((((((...((......))....))))))))).(((.......))).)))))....))))))))))...... ( -25.10) >DroSim_CAF1 22335 120 + 1 ACGCAAAUACACAUGCAAAGUGCCGCUGUUAAAUGGCUAAAUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUU ..(((........)))........((((((...(((((.......(((((((((...((......))....))))))))).(((.......))).)))))....)))))).......... ( -25.00) >DroEre_CAF1 24073 120 + 1 ACGCAAAUACACAUGCAAAGUGCCGCUGUUAAAUGACUGAAUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUU ..((......(((.((........)))))....((((((......(((((((((...((......))....))))))))).(....)..)))))).))(((((.....)))))....... ( -20.10) >DroYak_CAF1 24029 120 + 1 ACGCAAAUACACAUGCAAUGUGCCGCUGUUAAAUGGCUGAAUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUU ...(((...(((((...)))))..((((((...(((((.......(((((((((...((......))....))))))))).(((.......))).)))))....)))))))))....... ( -25.20) >consensus ACGCAAAUACACAUGCAAAGUGCCGCUGUUAAAUGGCUAAAUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUU ...(((...(((.......)))..((((((...(((((.......(((((((((...((......))....)))))))))....(((....))).)))))....)))))))))....... (-21.42 = -21.82 + 0.40)

| Location | 18,731,217 – 18,731,337 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18731217 120 - 22407834 AAUUUAUCAAACUGUUUAUUCGGCUAAACUGUCGUUGUCUCGUUUAGUUAAAAGGUAUAAGCCAUAAACUAAACGUUUAUUUAGCCAUUUAACAGGGGCACUUUGCUUGUGUAUUUGCGU ...........(((((.....((((((((....)))....(((((((((....(((....)))...)))))))))......)))))....)))))..(((...(((....)))..))).. ( -26.60) >DroSec_CAF1 23845 120 - 1 AAUUUAUCAAACUGUUUAUUCGGCUUAACUGUUGUUGUCUCGUUUAGUUAAAAGGUAUAAGCCAUAAACUAAACGUUUAUUUAGCCAUUUAACAGCGGCACUUUGCAUGUGUAUUUGAGU ......((((((((((.....((((((((....))))...(((((((((....(((....)))...))))))))).......))))....)))))..((((.......)))).))))).. ( -27.10) >DroSim_CAF1 22335 120 - 1 AAUUUAUCAAACUGUUUAUUCGGCUUAACUGUUGUUGUCUCGUUUAGUUAAAAGGUAUAAGCCAUAAACUAAACGUUUAUUUAGCCAUUUAACAGCGGCACUUUGCAUGUGUAUUUGCGU ...........(((((.....((((((((....))))...(((((((((....(((....)))...))))))))).......))))....)))))..(((...(((....)))..))).. ( -24.90) >DroEre_CAF1 24073 120 - 1 AAUUUAUCAAACUGUUUAUUCGGCUUAACUGUUGUUGUCUCGUUUAGUUAAAAGGUAUAAGCCAUAAACUAAACGUUUAUUCAGUCAUUUAACAGCGGCACUUUGCAUGUGUAUUUGCGU ..........((((..((..(((..((((....))))..))((((((((....(((....)))...)))))))))..))..)))).........(((((((.......))))...))).. ( -22.80) >DroYak_CAF1 24029 120 - 1 AAUUUAUCAAACUGUUUAUUCGGCUUAACUGUUGUUGUCUCGUUUAGUUAAAAGGUAUAAGCCAUAAACUAAACGUUUAUUCAGCCAUUUAACAGCGGCACAUUGCAUGUGUAUUUGCGU .......(((((((((.....((((((((....))))...(((((((((....(((....)))...))))))))).......))))....)))))..((((((...)))))).))))... ( -25.60) >consensus AAUUUAUCAAACUGUUUAUUCGGCUUAACUGUUGUUGUCUCGUUUAGUUAAAAGGUAUAAGCCAUAAACUAAACGUUUAUUUAGCCAUUUAACAGCGGCACUUUGCAUGUGUAUUUGCGU .......(((((((((.....((((((((....))))...(((((((((....(((....)))...))))))))).......))))....)))))..((((.......)))).))))... (-23.98 = -24.02 + 0.04)

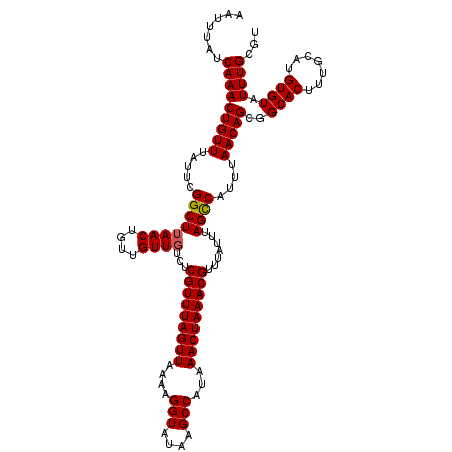
| Location | 18,731,257 – 18,731,377 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.93 |
| Mean single sequence MFE | -19.07 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18731257 120 + 22407834 AUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACGACAGUUUAGCCGAAUAAACAGUUUGAUAAAUUUAUGCGAGCUGCGACAAAAAAAAAAAACGGAAAAAUAAAU .....(((((((((...((......))....)))))))))......((.((((((.(((((((.....)))))(((....)))))))))))))........................... ( -19.90) >DroSec_CAF1 23885 116 + 1 AUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUUUAUGCGAGCUGCGACAAAA----AAAACGGAAAAAUGAAU .....(((((((((...((......))....))))))))).........((......(((......((((((((((....))).)))))))........----....))).....))... ( -18.87) >DroSim_CAF1 22375 118 + 1 AUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUUUAUGCGAGCUGCGACAAAAA--AAAAACGGAAAAAUAAAU .....(((((((((...((......))....)))))))))...........(((.(((((.(((((............))))).)).)))..))).....--.................. ( -18.70) >DroEre_CAF1 24113 114 + 1 AUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUUUAUGCGAGCUGCGACAAA------AAACGAAAAAAUAAAU .....(((((((((...((......))....)))))))))...........(((.(((((.(((((............))))).)).)))..)))...------................ ( -18.70) >DroYak_CAF1 24069 114 + 1 AUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUUUAUGCGAGCUGCGACAAA------AAACGGAAAAAUAAAU .....(((((((((...((......))....))))))))).................(((......((((((((((....))).))))))).......------...))).......... ( -19.19) >consensus AUAAACGUUUAGUUUAUGGCUUAUACCUUUUAACUAAACGAGACAACAACAGUUAAGCCGAAUAAACAGUUUGAUAAAUUUAUGCGAGCUGCGACAAAA____AAAACGGAAAAAUAAAU .....(((((((((...((......))....)))))))))..........................((((((((((....))).)))))))............................. (-18.10 = -18.10 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:55 2006