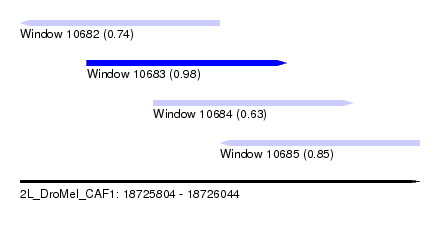
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,725,804 – 18,726,044 |
| Length | 240 |
| Max. P | 0.983063 |

| Location | 18,725,804 – 18,725,924 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.63 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -37.97 |
| Energy contribution | -38.61 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18725804 120 - 22407834 GCAUUGGGGCGAUUAGUAGCGCCUUUCGAUUUUUUUUUUCGCCCCUCGUUUUGGAGUGAAUGUUUUGGGCGCGUCCAUCGCAGAGGAGGAGUAACGAGGGGUGGGGGCAAAUUAAGCAGG ((...((((((........))))))..(((((...((..(((((((((((..........(((..((((....))))..)))..........)))))))))))..)).)))))..))... ( -40.85) >DroSec_CAF1 18390 118 - 1 GCAUUGGGGCGAUUAGUAGCGCCUUUCGAUUUU--UUUUCGCCCCUCGUUUUGGAGUGAAUGUUUUGGGCGCGUCCAUCGCAGAGGAGGAGUAACGAGGGGUGGGGGCAAAUUAAGCAGG ((...((((((........))))))..(((((.--((..(((((((((((..........(((..((((....))))..)))..........)))))))))))..)).)))))..))... ( -43.25) >DroSim_CAF1 16854 118 - 1 GCAUUGGGGCGAUUAGUAGCGCCUUUCGAUUUU--UUUUCGCCCCUCGUUUUGGAGUGAAUGUUUUGGGCGCGUCCAUCGCAGAGGAGGAGUAACGAGGGGUGGGGGCAAAUUAAGCAGG ((...((((((........))))))..(((((.--((..(((((((((((..........(((..((((....))))..)))..........)))))))))))..)).)))))..))... ( -43.25) >DroEre_CAF1 18512 117 - 1 GCAUUGGGGCGAUUAGUAGCGCCUUUCGAUUUU---UUUCGCCCCGCGUUUUGGAGUGAAUGUUUUGGGCGCGUCCAGUGCAGAGGAGGAGUAACGAGGGGUGGGGGCAAAUUAAGCAGG ((...((((((........))))))..((((((---(..((((((.((((..........(((.(((((....))))).)))..........)))).))))))..)).)))))..))... ( -38.65) >DroYak_CAF1 18546 117 - 1 GCAUUGGGGCGAUUAGUAGCGCCUUUCGAUUUU---UUUCGCCCCGUGUUUUGGAGUGAAUGUUUUGGGCGCGUCCAGUGCAGAGGCGGAGUAACGAGGGGUGGGGGCAAAUUAAGCAGG ((...((((((........))))))..((((((---(..((((((.((((.....((...(((.(((((....))))).)))...)).....)))).))))))..)).)))))..))... ( -38.10) >consensus GCAUUGGGGCGAUUAGUAGCGCCUUUCGAUUUU__UUUUCGCCCCUCGUUUUGGAGUGAAUGUUUUGGGCGCGUCCAUCGCAGAGGAGGAGUAACGAGGGGUGGGGGCAAAUUAAGCAGG ((...((((((........))))))..(((((...((..(((((((((((..........(((..((((....))))..)))..........)))))))))))..)).)))))..))... (-37.97 = -38.61 + 0.64)

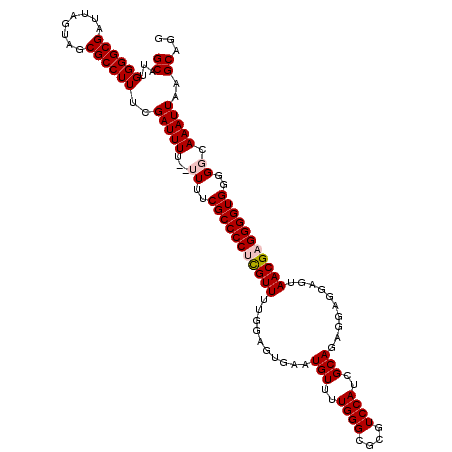
| Location | 18,725,844 – 18,725,964 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.84 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -26.67 |
| Energy contribution | -26.71 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18725844 120 + 22407834 CGAUGGACGCGCCCAAAACAUUCACUCCAAAACGAGGGGCGAAAAAAAAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUAUGGAUUCAAUGUAUGUAUCUGCAUCUGACGGAUACAU ...(((......)))..(((((.(.((((......(((((((.........((....)).........)))))))........)))).).)))))((((((((((....).))))))))) ( -32.61) >DroSec_CAF1 18430 114 + 1 CGAUGGACGCGCCCAAAACAUUCACUCCAAAACGAGGGGCGAAAA--AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAU ((.(((......)))....................(((((((...--....((....)).........)))))))........))......(((----(((((((.....)))))))))) ( -31.19) >DroSim_CAF1 16894 114 + 1 CGAUGGACGCGCCCAAAACAUUCACUCCAAAACGAGGGGCGAAAA--AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAU ((.(((......)))....................(((((((...--....((....)).........)))))))........))......(((----(((((((.....)))))))))) ( -31.19) >DroEre_CAF1 18552 113 + 1 CACUGGACGCGCCCAAAACAUUCACUCCAAAACGCGGGGCGAAA---AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAU ..(((...(((.....................)))(((((((..---....((....)).........)))))))........))).....(((----(((((((.....)))))))))) ( -31.96) >DroYak_CAF1 18586 113 + 1 CACUGGACGCGCCCAAAACAUUCACUCCAAAACACGGGGCGAAA---AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAU ...(((......)))..........(((.......(((((((..---....((....)).........)))))))...(....))))....(((----(((((((.....)))))))))) ( -30.46) >consensus CGAUGGACGCGCCCAAAACAUUCACUCCAAAACGAGGGGCGAAAA__AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG____UAUCUGCAUCUGGCGGAUACAU ...(((......)))..........(((.......(((((((.........((....)).........)))))))...(((..(((((...........))))).....))))))..... (-26.67 = -26.71 + 0.04)

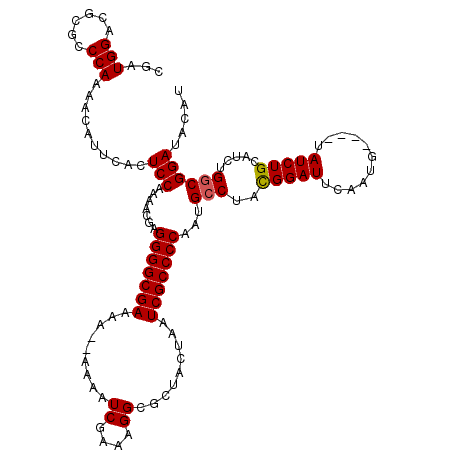
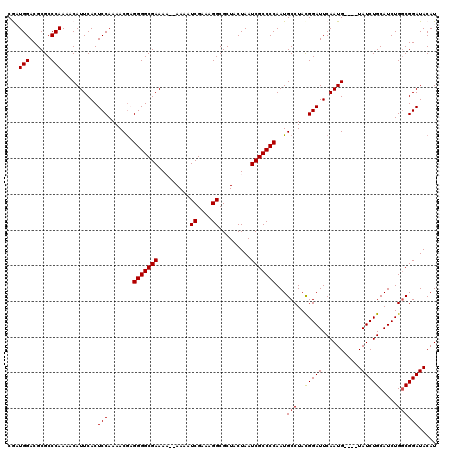
| Location | 18,725,884 – 18,726,004 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18725884 120 + 22407834 GAAAAAAAAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUAUGGAUUCAAUGUAUGUAUCUGCAUCUGACGGAUACAUGCGGCCGAUUUCCGUGCAUUUAACGCAUAAACGCUUUUUG ............(((((((((.....((((((((((.......))).......((((((((((((....).))))))))))))).)))))...((((.......))))...))))))))) ( -34.30) >DroSec_CAF1 18470 114 + 1 GAAAA--AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAUGCGGCCGAUUUCCGUGCAUUUAACGCAUAAACGCUUUUUG .....--.....(((((((((.....(((((((.(.....(....).....(((----(((((((.....))))))))))).)).)))))...((((.......))))...))))))))) ( -32.00) >DroSim_CAF1 16934 114 + 1 GAAAA--AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAUGCGGCCGAUUUCCGUGCAUUUAACGCAUAAACGCUUUUUG .....--.....(((((((((.....(((((((.(.....(....).....(((----(((((((.....))))))))))).)).)))))...((((.......))))...))))))))) ( -32.00) >DroEre_CAF1 18592 113 + 1 GAAA---AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAUGCGGCCGAUUUCCGUGCAUUUAACGCAUAAACGCUCUUUG ....---.....((((..(((.....(((((((.(.....(....).....(((----(((((((.....))))))))))).)).)))))...((((.......))))...)))..)))) ( -29.20) >DroYak_CAF1 18626 113 + 1 GAAA---AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG----UAUCUGCAUCUGGCGGAUACAUGCGGCCGAUUUCCGUGCAUUUAACGCAUAAACGCUUUUUG ....---.....(((((((((.....(((((((.(.....(....).....(((----(((((((.....))))))))))).)).)))))...((((.......))))...))))))))) ( -32.00) >consensus GAAAA__AAAAUCGAAAGGCGCUACUAAUCGCCCCAAUGCCUACGGAUUCAAUG____UAUCUGCAUCUGGCGGAUACAUGCGGCCGAUUUCCGUGCAUUUAACGCAUAAACGCUUUUUG ............((((((((((........))...(((((..(((((.((..((....(((((((.....)))))))....))...))..))))))))))............)))))))) (-25.20 = -25.44 + 0.24)

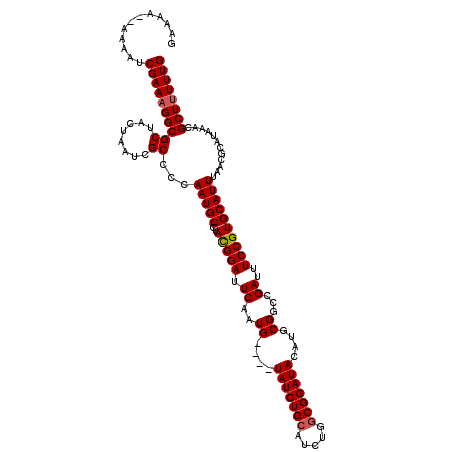
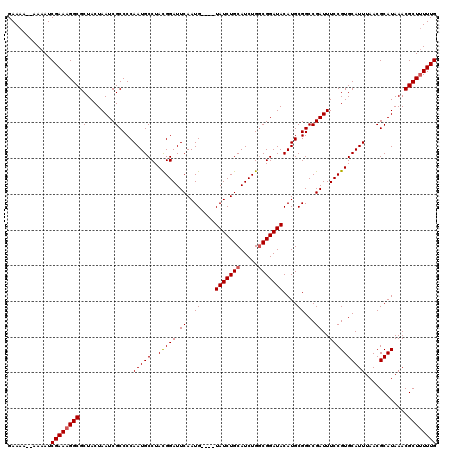
| Location | 18,725,924 – 18,726,044 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18725924 120 - 22407834 AGCGGCGAUAAUGAUAAUCCCCGAAUGGAAUAAAAGUGUCCAAAAAGCGUUUAUGCGUUAAAUGCACGGAAAUCGGCCGCAUGUAUCCGUCAGAUGCAGAUACAUACAUUGAAUCCAUAG .((((((((.......))).(((..((((.........))))....(((((((.....))))))).)))......)))))(((((((.((.....)).)))))))............... ( -31.00) >DroSec_CAF1 18508 116 - 1 AGCGGCGAUAAUGAUAAUCCCCGAACGGAAUAAAAGUGUCCAAAAAGCGUUUAUGCGUUAAAUGCACGGAAAUCGGCCGCAUGUAUCCGCCAGAUGCAGAUA----CAUUGAAUCCGUAG .((((((((.......))).(((...(((.........))).....(((((((.....))))))).)))......)))))(((((((.((.....)).))))----)))........... ( -31.40) >DroSim_CAF1 16972 116 - 1 AGCGGCGAUAAUGAUAAUCCCCGAACGGAAUAAAAGUGUCCAAAAAGCGUUUAUGCGUUAAAUGCACGGAAAUCGGCCGCAUGUAUCCGCCAGAUGCAGAUA----CAUUGAAUCCGUAG .((((((((.......))).(((...(((.........))).....(((((((.....))))))).)))......)))))(((((((.((.....)).))))----)))........... ( -31.40) >DroEre_CAF1 18629 116 - 1 AGCGGCGAUAAUGAUAAUCCCCGAACGGAAUAAAAGUGUCCAAAGAGCGUUUAUGCGUUAAAUGCACGGAAAUCGGCCGCAUGUAUCCGCCAGAUGCAGAUA----CAUUGAAUCCGUAG .((((((((.......))).(((...(((.........))).....(((((((.....))))))).)))......)))))(((((((.((.....)).))))----)))........... ( -31.40) >DroYak_CAF1 18663 116 - 1 AGCGGCGAUAAUGAUAAUCCCCGAACGGAAUAAAAGUGUCCAAAAAGCGUUUAUGCGUUAAAUGCACGGAAAUCGGCCGCAUGUAUCCGCCAGAUGCAGAUA----CAUUGAAUCCGUAG .((((((((.......))).(((...(((.........))).....(((((((.....))))))).)))......)))))(((((((.((.....)).))))----)))........... ( -31.40) >consensus AGCGGCGAUAAUGAUAAUCCCCGAACGGAAUAAAAGUGUCCAAAAAGCGUUUAUGCGUUAAAUGCACGGAAAUCGGCCGCAUGUAUCCGCCAGAUGCAGAUA____CAUUGAAUCCGUAG .((((((((.......))).(((...(((.........))).....(((((((.....))))))).)))......))))).((((((.....))))))...................... (-28.14 = -28.14 + 0.00)

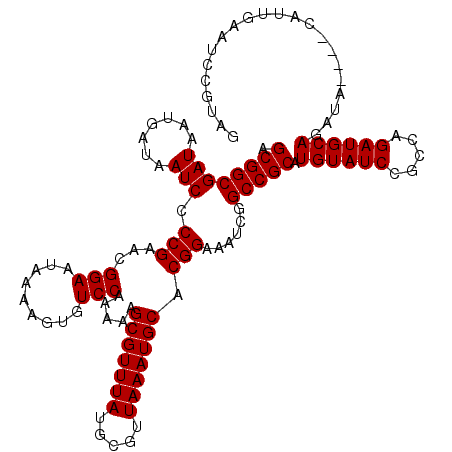
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:39 2006