
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,724,715 – 18,724,989 |
| Length | 274 |
| Max. P | 0.939052 |

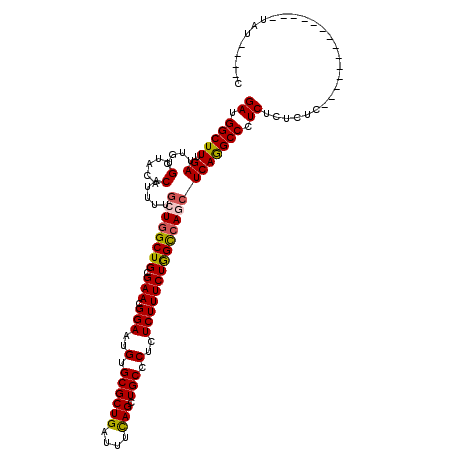
| Location | 18,724,715 – 18,724,831 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.55 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -24.70 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
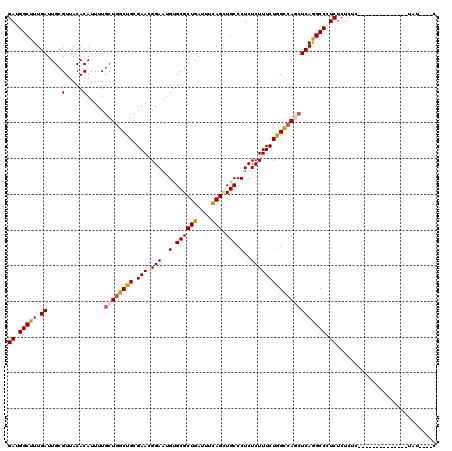
>2L_DroMel_CAF1 18724715 116 - 22407834 GAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAACGGAAUGUGCGCUGAUUUCAGCUGCCCUCUCUUUCUAGCCAGCUCAGGCCCUCUCUCUCUCUCUUUAUUUAUCUAU----C ((.(((((.((....(.....).....((((((((.(((.(((..(.((((((....)))).)).)..))))))))))))))))))))).)).......................----. ( -32.60) >DroSec_CAF1 17323 102 - 1 GAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAACGGAAUGUGCGCUGAUUUCAGUUGCCCUCUCUUUCUGGCCAGCUCAGGCCCUCUCUCUC--------------UAU----C ((.(((((.((....(.....).....((((((((.(((.(((..(.((((((....))).))).)..))))))))))))))))))))).))......--------------...----. ( -29.40) >DroSim_CAF1 15811 102 - 1 GAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAACGGAAUGUGCGCUGAUUUCAGUUGCCCUCUCUUUCUGGCCAGCUCAGGCCCUCUCUCUC--------------UAU----C ((.(((((.((....(.....).....((((((((.(((.(((..(.((((((....))).))).)..))))))))))))))))))))).))......--------------...----. ( -29.40) >DroEre_CAF1 17483 102 - 1 GAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAACGGAAUGCGCGCUGAUUUUAGCUGCCCUCUCUUUCUGGCCAAGUCAGGCCCUCUCUCUC--------------GCU----C ((.(((((.(((((((..........)))((((((.(((.(((..(((.((((....)))))))..))).))).))))))))))))))).))......--------------...----. ( -27.80) >DroYak_CAF1 17538 106 - 1 GAUGGCUUUGAUUGCGUUACACAUUUUGCUCGCUGCGAACGGAAUGUGCGCUGAUUUUAGAUGCCCUCUCUUUCUGGUCAUCUCAAGCCAUCUCUCUC--------------UGUAACAA ((((((((.((..(((...(((((((((.(((...))).))))))))))))........((((.((.........)).))))))))))))))......--------------........ ( -31.70) >consensus GAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAACGGAAUGUGCGCUGAUUUCAGCUGCCCUCUCUUUCUGGCCAGCUCAGGCCCUCUCUCUC______________UAU____C ((.(((((.((....(.....).....((((((((.(((.(((..(.((((((....))).))).)..))))))))))))))))))))).))............................ (-24.70 = -24.78 + 0.08)

| Location | 18,724,791 – 18,724,909 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -39.80 |
| Energy contribution | -39.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18724791 118 - 22407834 AUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GAGCGUAAUCCAAGGUAGUUAAUUGCGCCUUAAACCGGAAGCGAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAAC .((((((.......((.((.....)))).(..((((--((((((((((.(((((.....(((((.((.......))..))))).)))))))))))))......))))))..))))))).. ( -40.70) >DroSec_CAF1 17385 118 - 1 AUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GAGCGUAAUCCAAGGUAGUUAAUUGCGCCUUAAACCGGAAGCGAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAAC .((((((.......((.((.....)))).(..((((--((((((((((.(((((.....(((((.((.......))..))))).)))))))))))))......))))))..))))))).. ( -40.70) >DroSim_CAF1 15873 118 - 1 AUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GAGCGUAAUCCAAGGUAGUUAAUUGCGCCUUAAACCGGAAGCGAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAAC .((((((.......((.((.....)))).(..((((--((((((((((.(((((.....(((((.((.......))..))))).)))))))))))))......))))))..))))))).. ( -40.70) >DroEre_CAF1 17545 120 - 1 AUCGCAGGGGAGGAGGAGCUGCUUGCCCACUGGCAACAGAGCGUAAUCCAAGGUAGUUAAUUGCGCCUUAAACCGGAAGCGAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAAC .(((((((((..(((......))).))).(..((((...(((((((((.(((((.....(((((.((.......))..))))).)))))))))))))).......))))..))))))).. ( -41.60) >DroYak_CAF1 17604 118 - 1 AUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GAGCGUAAUCCAAGGUAGUUAAUUGCGCCUUAAACCGGAAGCGAUGGCUUUGAUUGCGUUACACAUUUUGCUCGCUGCGAAC .((((((.(.((.((((..((.(((((....)))))--.(((((((((.(((((.....(((((.((.......))..))))).)))))))))))))).))..)))).))).)))))).. ( -40.80) >consensus AUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA__GAGCGUAAUCCAAGGUAGUUAAUUGCGCCUUAAACCGGAAGCGAUGGCUUUGAUUGCGUUACACAUUUUGCUGGCUGCGAAC .((((((.......((.((.....)))).(..((((...(((((((((.(((((.....(((((.((.......))..))))).)))))))))))))).......))))..))))))).. (-39.80 = -39.80 + 0.00)

| Location | 18,724,871 – 18,724,989 |
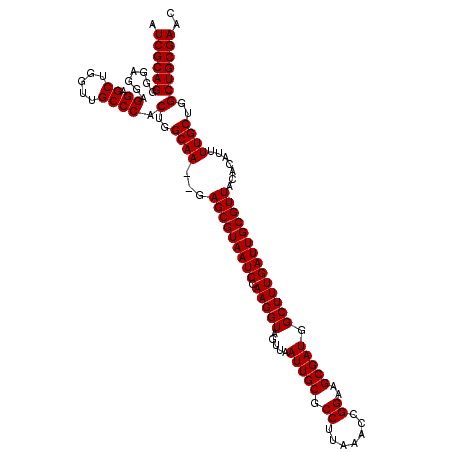
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -44.71 |
| Consensus MFE | -39.06 |
| Energy contribution | -39.74 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
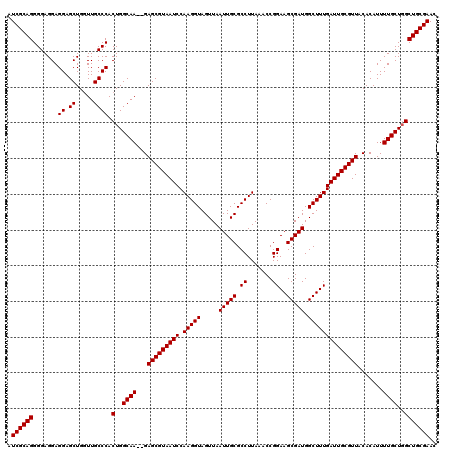
>2L_DroMel_CAF1 18724871 118 + 22407834 UC--UUGCCAGUGGGCAACCAGCUCCUCCUCCCCUGCGAUUCCCACUGCAAUUGGCGGUGGGAGCUUUGUAGUGGCGGUUCGGACUGGGGACCCGGCUUUGCUAUAUAUAGCGUCAUGGC ..--..((((..(((...((((.(((.((.((.((((((((((((((((.....))))))))))..)))))).)).))...)))))))...)))(((...((((....))))))).)))) ( -53.30) >DroSec_CAF1 17465 118 + 1 UC--UUGCCAGUGGGCAACCAGCUCCUCCUCCCCUGCGAUUCCCACUGCAUUUGGCGGUGGAAGCUUUGUAGUGCCCGUUCGGACUGGGGACCCGACUUUGCUAUAUUUAGCGUCAUGGC ..--..((((..(((...((((.(((......(((((((...(((((((.....))))))).....)))))).).......)))))))...)))(((...((((....))))))).)))) ( -41.02) >DroSim_CAF1 15953 118 + 1 UC--UUGCCAGUGGGCAACCAGCUCCUCCUCCCCUGCGAUUCCCACUGCAAUUGGCGGUGGGAGCUUUGUAGUGCCCGUUCGGACUGGGGACCCGACUUUGCUAUAUAUAGCGUCAUGGC ..--..((((..(((...((((.(((......(((((((((((((((((.....))))))))))..)))))).).......)))))))...)))(((...((((....))))))).)))) ( -46.02) >DroEre_CAF1 17625 118 + 1 UCUGUUGCCAGUGGGCAAGCAGCUCCUCCUCCCCUGCGAUUCCCACUGCAAUUGGCGGUGGGAGCUUUGUAGUGCCCGUUCGGACGG-GGACCCGAUUUU-CUAUAUAUAGCGUCAUGGC ......((((...(((..((((...........))))...(((((((((.....)))))))))(((.((((((.(((((....))))-).))..(.....-)..)))).)))))).)))) ( -42.60) >DroYak_CAF1 17684 117 + 1 UC--UUGCCAGUGGGCAACCAGCUCCUCCUCCCCUGCGAUUUCCACUGCAAUUGGCGGUGGGAGCUUUGUAGUGCCCGUUCGGACGGGGGACCCGAUUUU-CUAUAUAUAGCGUCAUGGC ..--..((((.(((....)))(((((((((((...(((..(((((((((.....)))))))))((........)).)))..))).))))))...(.....-)........))....)))) ( -40.60) >consensus UC__UUGCCAGUGGGCAACCAGCUCCUCCUCCCCUGCGAUUCCCACUGCAAUUGGCGGUGGGAGCUUUGUAGUGCCCGUUCGGACUGGGGACCCGACUUUGCUAUAUAUAGCGUCAUGGC ......((((..(((...((((.(((......(((((((((((((((((.....))))))))))..)))))).).......)))))))...)))(((...((((....))))))).)))) (-39.06 = -39.74 + 0.68)

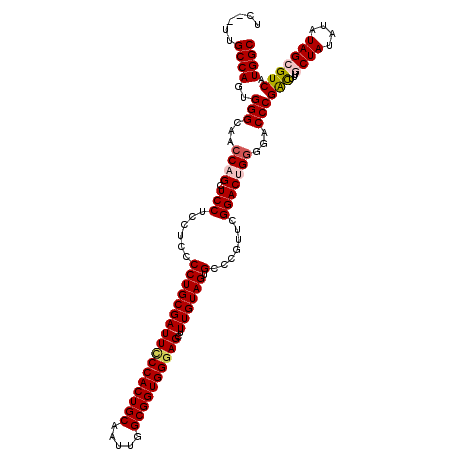
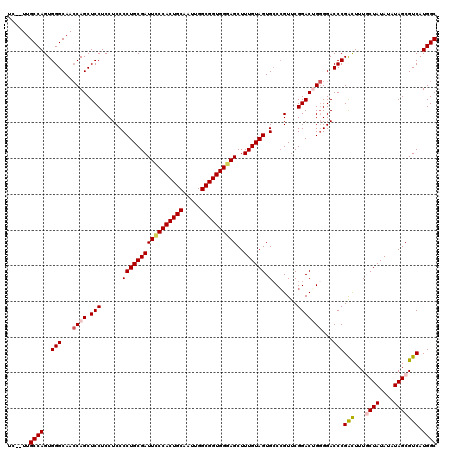
| Location | 18,724,871 – 18,724,989 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -37.54 |
| Energy contribution | -38.38 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18724871 118 - 22407834 GCCAUGACGCUAUAUAUAGCAAAGCCGGGUCCCCAGUCCGAACCGCCACUACAAAGCUCCCACCGCCAAUUGCAGUGGGAAUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GA ((((....((((....))))......((((..(((((((...((.((.((.......((((((.((.....)).))))))......)))).)).))).))))..))))..))))..--.. ( -45.22) >DroSec_CAF1 17465 118 - 1 GCCAUGACGCUAAAUAUAGCAAAGUCGGGUCCCCAGUCCGAACGGGCACUACAAAGCUUCCACCGCCAAAUGCAGUGGGAAUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GA ((((.(((((((....))))...)))((((..((.((((....)))).......((((.((.((.((...(((.((....)).)))..)).)).))))))))..))))..))))..--.. ( -44.30) >DroSim_CAF1 15953 118 - 1 GCCAUGACGCUAUAUAUAGCAAAGUCGGGUCCCCAGUCCGAACGGGCACUACAAAGCUCCCACCGCCAAUUGCAGUGGGAAUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GA ((((.(((((((....))))...)))((((..((.((((....)))).......((((((..((.((..((((.((....)).)))).)).)).))))))))..))))..))))..--.. ( -45.30) >DroEre_CAF1 17625 118 - 1 GCCAUGACGCUAUAUAUAG-AAAAUCGGGUCC-CCGUCCGAACGGGCACUACAAAGCUCCCACCGCCAAUUGCAGUGGGAAUCGCAGGGGAGGAGGAGCUGCUUGCCCACUGGCAACAGA .((.(((..(((....)))-....)))..(((-((((((....))))........((((((((.((.....)).))))))...)).))))))).((.((.....)))).(((....))). ( -40.30) >DroYak_CAF1 17684 117 - 1 GCCAUGACGCUAUAUAUAG-AAAAUCGGGUCCCCCGUCCGAACGGGCACUACAAAGCUCCCACCGCCAAUUGCAGUGGAAAUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA--GA ((((.((..(((....)))-....))((((..((.((((....)))).......((((((..((.((..((((.((....)).)))).)).)).))))))))..))))..))))..--.. ( -41.00) >consensus GCCAUGACGCUAUAUAUAGCAAAGUCGGGUCCCCAGUCCGAACGGGCACUACAAAGCUCCCACCGCCAAUUGCAGUGGGAAUCGCAGGGGAGGAGGAGCUGGUUGCCCACUGGCAA__GA ((((....((((....))))......((((..((.((((....)))).......((((((..((.((...(((.((....)).)))..)).)).))))))))..))))..))))...... (-37.54 = -38.38 + 0.84)

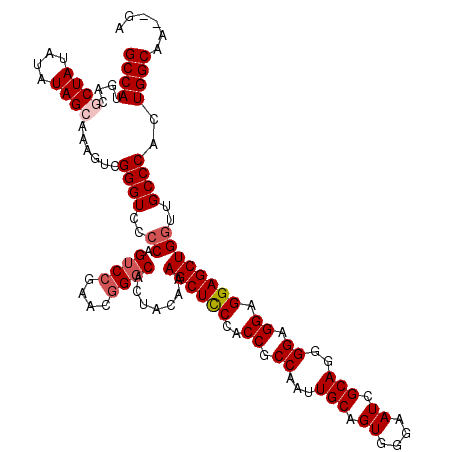

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:33 2006