
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,721,765 – 18,721,962 |
| Length | 197 |
| Max. P | 0.998824 |

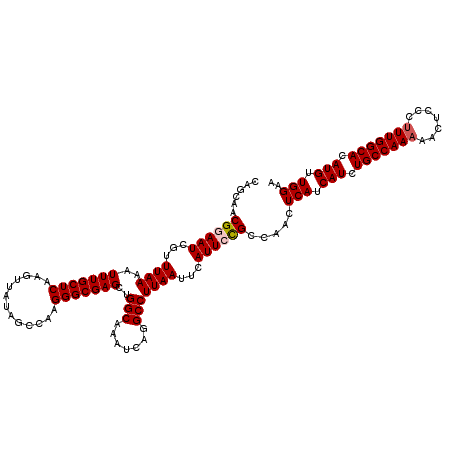
| Location | 18,721,765 – 18,721,885 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18721765 120 + 22407834 CAGCAACGGAAUCGUUUAAAAUUUGCUCAAGUUAUAGCCAAGGGCGAGCUGGCAAAUCAGGCCUUAAUUCAUUCCGCCAACUCAUCAUCUGCCAAAAACUCCCGUUGGCACAUGUUGGAA (((((..(((((...((((.(((((((..((((...(((...))).)))))))))))......))))...)))))((((((......................))))))...)))))... ( -28.25) >DroSec_CAF1 14439 120 + 1 CAGCAACGGAAUCGUUUAAAAUUUGCUCAAGUUAUAGGCAAGGGCGAGCUGGCAAAUCAGGCCUUAAUUCAUUCCGCCAACUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAA (((((.((((((.(((((.(((((....))))).))))).(((((...((((....))))))))).....)))))).............(((((((.......)))))))..)))))... ( -29.80) >DroSim_CAF1 12915 120 + 1 CAGCAACGGAAUCGUUUAAAAUUUGCUCAAGUUAUAGCCAAGGGCGAGCUGGCAAAUCAGGCCUUAAUUCAUUCCGCCAACUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAA (((((.((((((...((((.(((((((..((((...(((...))).)))))))))))......))))...)))))).............(((((((.......)))))))..)))))... ( -29.00) >DroEre_CAF1 14621 119 + 1 CACCAACG-AAUCGUUUAAAAUUUGCUCAAGCUAUAGCCAAGGGCGAGCUGGCAAAUCAGGCCUUAAUUCAUUCCGCCAACUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAA ..((((((-(..........(((((((..((((...(((...))).)))))))))))..(((.............)))......))...(((((((.......)))))))...))))).. ( -31.72) >DroYak_CAF1 14682 119 + 1 CACCAACG-AAUCGUUUAAAAUUUGCUCAACUUAAAGCCAAGGGCGAGCUGGCAAAUCAGGCCUUAAUUCAUUCUGCCUACUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAA ..((((((-....((.........))..........((((((((.(((.(((((....((((.............))))..........)))))....)))))))))))...)))))).. ( -29.96) >consensus CAGCAACGGAAUCGUUUAAAAUUUGCUCAAGUUAUAGCCAAGGGCGAGCUGGCAAAUCAGGCCUUAAUUCAUUCCGCCAACUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAA ......((((((...((((..(((((((.............)))))))..(((.......)))))))...)))))).....(((.(((.(((((((.......))))))).))).))).. (-24.52 = -24.96 + 0.44)


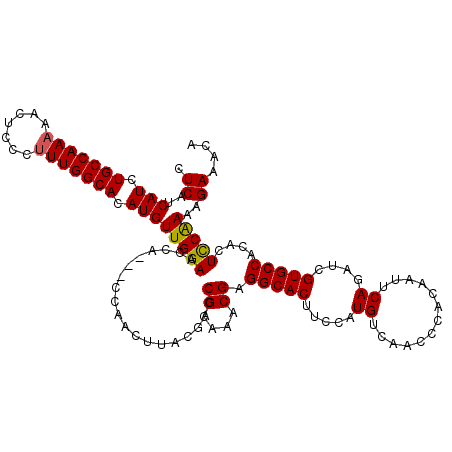
| Location | 18,721,845 – 18,721,962 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -20.10 |
| Energy contribution | -19.98 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18721845 117 + 22407834 CUCAUCAUCUGCCAAAAACUCCCGUUGGCACAUGUUGGAAUCA---CCAACUUACGACCGCAAACGAGGCACUUCCAUGUCAACCCACAAUUCAGAUCGUGCCACACUCCAAAAGAAACA .((..(((.((((((.........)))))).)))(((((....---........((........)).(((((.((..(((......))).....))..)))))....)))))..)).... ( -19.90) >DroSec_CAF1 14519 117 + 1 CUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAACCA---CCAGCUUACGACCGCAAACGAGGCACUUCCAUGUCAACCCACAAUUCAGAUCGUGCCACACUCCAAAAGAAACA .((..(((.(((((((.......))))))).)))(((((....---...((........))......(((((.((..(((......))).....))..)))))....)))))..)).... ( -23.10) >DroSim_CAF1 12995 117 + 1 CUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAACCA---CCAGCUUACGACCGCAAACGAGGCACUUCCAUGUCAACCCACAAUUCAGAUCGUGCCACACUCCAAAAGAAACA .((..(((.(((((((.......))))))).)))(((((....---...((........))......(((((.((..(((......))).....))..)))))....)))))..)).... ( -23.10) >DroEre_CAF1 14700 120 + 1 CUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAACCACCACCAACUUAUGACCGCAAACGAGGCACUUUCAUGUCAACCCACAAUUCAGUUCGUGCCACACUCCGAAAGAAACA ....((((.(((((((.......)))))))...(((((........)))))..))))..........(((((.....((.............))....)))))......(....)..... ( -21.92) >DroYak_CAF1 14761 117 + 1 CUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAACCA---CCAACUUAUGACCGCAAACGAGGCACUUUCGUGUCAACCCACAAUUCAGAUCGUGCCACACUUCAAAAGAAACA .(((.(((.(((((((.......))))))).))).))).....---....(((.(((.((....)).(((((..((.(((......))).....))..))))).....))).)))..... ( -24.90) >consensus CUCAUCAUCUGCCAAAAACUCCCUUUGGCACAUGUUGGAACCA___CCAACUUACGACCGCAAACGAGGCACUUCCAUGUCAACCCACAAUUCAGAUCGUGCCACACUCCAAAAGAAACA .((..(((.(((((((.......))))))).)))(((((...................((....)).(((((.....((.............))....)))))....)))))..)).... (-20.10 = -19.98 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:23 2006