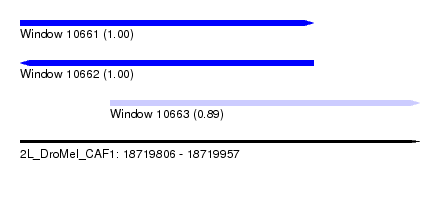
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,719,806 – 18,719,957 |
| Length | 151 |
| Max. P | 0.999894 |

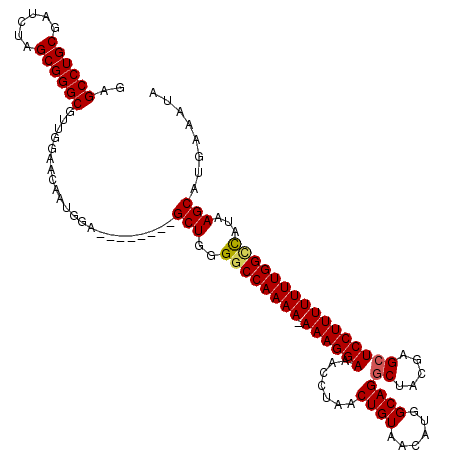
| Location | 18,719,806 – 18,719,917 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -36.90 |
| Energy contribution | -37.02 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18719806 111 + 22407834 GAGCCUGCGAUCUAGCGGGCGUUGGAACAAUGGA--------GCUGGGGCCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUA ..((((((......))))))..............--------(((..((((((((-((((((.......((((......))))((.....))))))))))))))))...)))........ ( -44.40) >DroSec_CAF1 12527 111 + 1 GAGCCUGCGAUCUAGCGGGCGUUGGAACAAUGGA--------GCUGGGGCCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUA ..((((((......))))))..............--------(((..((((((((-((((((.......((((......))))((.....))))))))))))))))...)))........ ( -44.40) >DroSim_CAF1 10962 112 + 1 GAGCCUGCGAUCUAGCGGGCGUUGGAACAAUGGA--------GCUGGGGCCAAAAAAAAGGAAACCUAACUGUAACUUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUA ..((((((......))))))..............--------(((..(((((((((((.(((.......((((......))))((.....))))))))))))))))...)))........ ( -40.60) >DroEre_CAF1 12690 111 + 1 GAGCCUGCGAUCUAGCGGGCGUUGGAACAAUGGA--------GCUGGGACCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUA ..((((((......))))))..............--------(((.((.((((((-((((((.......((((......))))((.....))))))))))))))))...)))........ ( -40.20) >DroYak_CAF1 12688 119 + 1 GAGCCUGCGAUCUAGCGGGCGUUGGAACAAUGGAGCUGUGGAGCUGGGGCCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGCCUACGAGCUCCUUUUUUUGGUUAUAAGCAUGAAAUA ..((((((......))))))(((..(((...((((((((((.((((((.((....-...))...))...(........).)))))))).))))))........)))...)))........ ( -37.70) >consensus GAGCCUGCGAUCUAGCGGGCGUUGGAACAAUGGA________GCUGGGGCCAAAA_AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUA ..((((((......))))))......................(((..((((((((.((((((.......((((......))))((.....))))))))))))))))...)))........ (-36.90 = -37.02 + 0.12)

| Location | 18,719,806 – 18,719,917 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18719806 111 - 22407834 UAUUUCAUGCUUAUGGCCAAAAAAAGGAGCUCGUAGCCUGCCAUGUUACAGUUAGGUUUCCUUU-UUUUGGCCCCAGC--------UCCAUUGUUCCAACGCCCGCUAGAUCGCAGGCUC ........(((...(((((((((((((((..(.((((.((........)))))).).)))))))-))))))))..)))--------..............(((.((......)).))).. ( -36.00) >DroSec_CAF1 12527 111 - 1 UAUUUCAUGCUUAUGGCCAAAAAAAGGAGCUCGUAGCCUGCCAUGUUACAGUUAGGUUUCCUUU-UUUUGGCCCCAGC--------UCCAUUGUUCCAACGCCCGCUAGAUCGCAGGCUC ........(((...(((((((((((((((..(.((((.((........)))))).).)))))))-))))))))..)))--------..............(((.((......)).))).. ( -36.00) >DroSim_CAF1 10962 112 - 1 UAUUUCAUGCUUAUGGCCAAAAAAAGGAGCUCGUAGCCUGCCAAGUUACAGUUAGGUUUCCUUUUUUUUGGCCCCAGC--------UCCAUUGUUCCAACGCCCGCUAGAUCGCAGGCUC ........(((...(((((((((((((((..(.((((.((........)))))).).)))).)))))))))))..)))--------..............(((.((......)).))).. ( -32.20) >DroEre_CAF1 12690 111 - 1 UAUUUCAUGCUUAUGGCCAAAAAAAGGAGCUCGUAGCCUGCCAUGUUACAGUUAGGUUUCCUUU-UUUUGGUCCCAGC--------UCCAUUGUUCCAACGCCCGCUAGAUCGCAGGCUC ........(((...(((((((((((((((..(.((((.((........)))))).).)))))))-))))))))..)))--------..............(((.((......)).))).. ( -33.30) >DroYak_CAF1 12688 119 - 1 UAUUUCAUGCUUAUAACCAAAAAAAGGAGCUCGUAGGCUGCCAUGUUACAGUUAGGUUUCCUUU-UUUUGGCCCCAGCUCCACAGCUCCAUUGUUCCAACGCCCGCUAGAUCGCAGGCUC ........(((.....(((((((((((((..(...(((((........))))).)..)))))))-))))))....)))......................(((.((......)).))).. ( -29.80) >consensus UAUUUCAUGCUUAUGGCCAAAAAAAGGAGCUCGUAGCCUGCCAUGUUACAGUUAGGUUUCCUUU_UUUUGGCCCCAGC________UCCAUUGUUCCAACGCCCGCUAGAUCGCAGGCUC ........(((...(((((((((((((((..(.(((.(((........)))))).).))))))).))))))))..)))......................(((.((......)).))).. (-28.90 = -29.18 + 0.28)



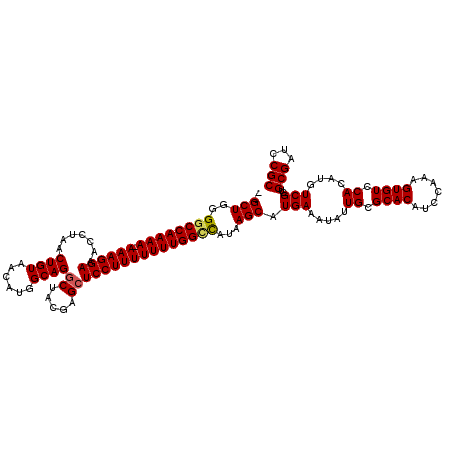
| Location | 18,719,840 – 18,719,957 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18719840 117 + 22407834 --GCUGGGGCCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUAUUGCGCACAUCCAAAGUGUCCACAUGUCGCUGCGAUCCGC --((.((((((((((-((((((.......((((......))))((.....))))))))))))))))....(((.(..((((((.((((.......)))).)).))))..))))...)))) ( -38.70) >DroSec_CAF1 12561 117 + 1 --GCUGGGGCCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUAUUGCGCACAUCCAAAGUGUCCACAUGUCGCUGCGAUCCGC --((.((((((((((-((((((.......((((......))))((.....))))))))))))))))....(((.(..((((((.((((.......)))).)).))))..))))...)))) ( -38.70) >DroSim_CAF1 10996 118 + 1 --GCUGGGGCCAAAAAAAAGGAAACCUAACUGUAACUUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUAUUGCGCACAUCCAAAGUGUCCACAUGUCGCUGCGAUCCGC --((.(((((((((((((.(((.......((((......))))((.....))))))))))))))))....(((.(..((((((.((((.......)))).)).))))..))))...)))) ( -34.90) >DroEre_CAF1 12724 117 + 1 --GCUGGGACCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUAUUGCGCACAUCCAAAGUGUCCACAUGUCGGAGCGAUCCGC --(((.((.((((((-((((((.......((((......))))((.....))))))))))))))))...))).....((((((.((((.......)))).)).))))((((....)))). ( -35.70) >DroYak_CAF1 12728 119 + 1 GAGCUGGGGCCAAAA-AAAGGAAACCUAACUGUAACAUGGCAGCCUACGAGCUCCUUUUUUUGGUUAUAAGCAUGAAAUAUUGCGCACAUCCAAAGUGUCCACAUGUCGGAGCGAUCCGC ..(((..((((((((-((((((.......((((......)))).........))))))))))))))...))).....((((((.((((.......)))).)).))))((((....)))). ( -32.69) >consensus __GCUGGGGCCAAAA_AAAGGAAACCUAACUGUAACAUGGCAGGCUACGAGCUCCUUUUUUUGGCCAUAAGCAUGAAAUAUUGCGCACAUCCAAAGUGUCCACAUGUCGCUGCGAUCCGC ..(((..((((((((.((((((.......((((......))))((.....))))))))))))))))...))).(((.....((.((((.......)))).))....)))..(((...))) (-29.20 = -29.32 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:19 2006