
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,717,001 – 18,717,142 |
| Length | 141 |
| Max. P | 0.843260 |

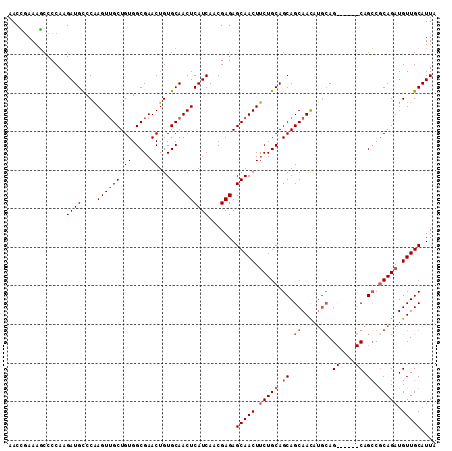
| Location | 18,717,001 – 18,717,102 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -23.58 |
| Energy contribution | -25.20 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18717001 101 + 22407834 AACCGAAAGCCCCAAGAUGCCCAAGUUGCUGUGGCGAACUGUGCAACUCAUCAACGAGAGCAACUUCUGCAGCAGCAAU----AG------CAGCCGCAGAUGUUGCAUUA ...(....)......((((....((((((.((.....))...)))))))))).......(((((.(((((.((.((...----.)------).)).))))).))))).... ( -31.70) >DroSec_CAF1 9784 108 + 1 AACCGAAAGCCCCAAGAUGCCCAAGUUGCUGUGGCGAACUGUGCAACUCAUCAACGAGAGCAACUUCUGCAGCAGCAACAUGCAGCA---GCAGCCGCAGAUGUUGCAUUA ...(....)......(((((....((((((((.(((((...(((..(((......))).)))..))).)).)))))))).(((.((.---...)).)))......))))). ( -32.30) >DroSim_CAF1 8215 105 + 1 AACUGAAAGCCCCAAGAUGCCCAAGUUGCUGUGGCGAACUGUGCAACUCAUCAACGAGAGCAACUUCUGCAGCAGCAACAUGCAG------CAGCCGCAGAUGUUGCAUUA ........................((((((((.(((((...(((..(((......))).)))..))).)).))))))))((((((------((........)))))))).. ( -31.70) >DroEre_CAF1 9929 111 + 1 AACGAAAACCCCCAAGAUGUCCGAGUGGCUGUGGCGAACUGUGCAACUCAUCAACGAGAGCAACUCCUGCAGCAGCAACAUGCAGCCGCAGCAGGCCCAGAUGUUGCAUUA ...............((((((.(.((.((((((((......(((..(((......))).(((.....))).)))((.....)).))))))))..)).).))))))...... ( -31.70) >consensus AACCGAAAGCCCCAAGAUGCCCAAGUUGCUGUGGCGAACUGUGCAACUCAUCAACGAGAGCAACUUCUGCAGCAGCAACAUGCAG______CAGCCGCAGAUGUUGCAUUA ...............((((....((((((.((.....))...)))))))))).......(((((.(((((.((.((.....)).((....)).)).))))).))))).... (-23.58 = -25.20 + 1.62)


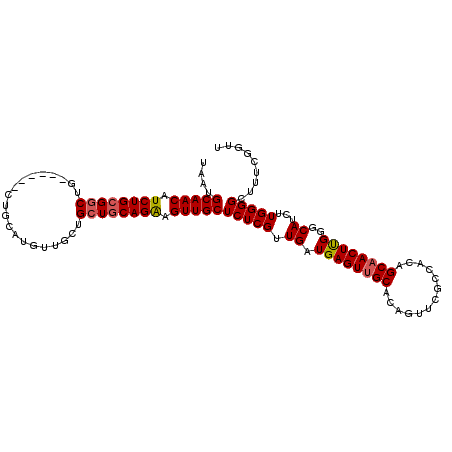
| Location | 18,717,001 – 18,717,102 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -37.25 |
| Consensus MFE | -28.27 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18717001 101 - 22407834 UAAUGCAACAUCUGCGGCUG------CU----AUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCACAGUUCGCCACAGCAACUUGGGCAUCUUGGGGCUUUCGGUU ....(((((.((((((((.(------(.----...)).)))))))).))))).....((((.(((((.((.((...(((.(((....))))))..)))).))))))))).. ( -37.00) >DroSec_CAF1 9784 108 - 1 UAAUGCAACAUCUGCGGCUGC---UGCUGCAUGUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCACAGUUCGCCACAGCAACUUGGGCAUCUUGGGGCUUUCGGUU ....(((((.((((((((.((---.((.....)).)).)))))))).))))).....((((.(((((.((.((...(((.(((....))))))..)))).))))))))).. ( -38.40) >DroSim_CAF1 8215 105 - 1 UAAUGCAACAUCUGCGGCUG------CUGCAUGUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCACAGUUCGCCACAGCAACUUGGGCAUCUUGGGGCUUUCAGUU ....(((((.((((((((.(------(........)).)))))))).))))).....((((.(((((.((.((...(((.(((....))))))..)))).))))))))).. ( -37.80) >DroEre_CAF1 9929 111 - 1 UAAUGCAACAUCUGGGCCUGCUGCGGCUGCAUGUUGCUGCUGCAGGAGUUGCUCUCGUUGAUGAGUUGCACAGUUCGCCACAGCCACUCGGACAUCUUGGGGGUUUUCGUU ...(((((((((.((((((.(((((((.((.....)).))))))).))..)))).....)))..))))))...........((((.(((((.....)))))))))...... ( -35.80) >consensus UAAUGCAACAUCUGCGGCUG______CUGCAUGUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCACAGUUCGCCACAGCAACUUGGGCAUCUUGGGGCUUUCGGUU ....(((((.((((((((....................)))))))).)))))(((((.((.((((((((.............))))))))..))...)))))......... (-28.27 = -28.64 + 0.37)

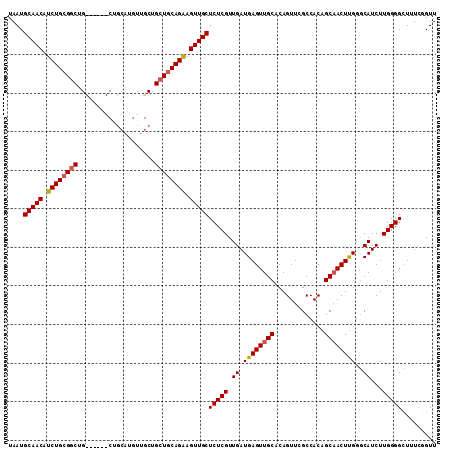
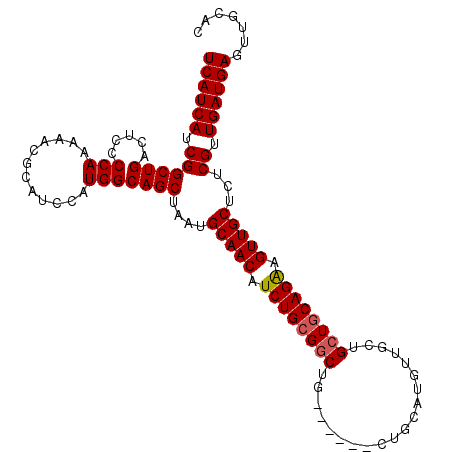
| Location | 18,717,041 – 18,717,142 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -29.64 |
| Energy contribution | -29.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18717041 101 - 22407834 UCAUCAUCGGCUGACUCCCCAAAAACGCAUCCAUGGCAGCUAAUGCAACAUCUGCGGCUG------CU----AUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCAC ((((((.((((((.....(((............)))))))....(((((.((((((((.(------(.----...)).)))))))).)))))...)).))))))....... ( -37.40) >DroSec_CAF1 9824 108 - 1 UCAUCAUCGGCUGACUCCCCAAAAACGCAUCCAUGGCAGCUAAUGCAACAUCUGCGGCUGC---UGCUGCAUGUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCAC ((((((.((((((.....(((............)))))))....(((((.((((((((.((---.((.....)).)).)))))))).)))))...)).))))))....... ( -38.80) >DroSim_CAF1 8255 105 - 1 UCAUCAUCGGCUGACUCCCCAAAAACGCAUCCAUGGCAGCUAAUGCAACAUCUGCGGCUG------CUGCAUGUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCAC ((((((.((((((.....(((............)))))))....(((((.((((((((.(------(........)).)))))))).)))))...)).))))))....... ( -37.40) >DroEre_CAF1 9969 111 - 1 UCAUCAUCGGCUGACUCCCCAAAAACGCAUCCAUGGCAGCUAAUGCAACAUCUGGGCCUGCUGCGGCUGCAUGUUGCUGCUGCAGGAGUUGCUCUCGUUGAUGAGUUGCAC ((((((.((((.((((((.........((....))(((((....(((((((...((((......))))..))))))).))))).))))))))...)).))))))....... ( -39.30) >consensus UCAUCAUCGGCUGACUCCCCAAAAACGCAUCCAUGGCAGCUAAUGCAACAUCUGCGGCUG______CUGCAUGUUGCUGCUGCAGAAGUUGCUCUCGUUGAUGAGUUGCAC ((((((.((((((.....(((............)))))))....(((((.((((((((....................)))))))).)))))...)).))))))....... (-29.64 = -29.95 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:12 2006